Climate scenario data#
In this notebook we will…
… aggregate and download climate scenario data from the Coupled Model Intercomparison Project Phase 6 (CMIP6) for our catchment,
… preprocess the data,
… compare the CMIP6 models with our reanalysis data and adjust them for biases,
… and visualize the data before and after bias adjustment.
The NEX-GDDP-CMIP6 dataset we are going to use has been downscaled to 27830 m resolution by the NASA Climate Analytics Group and is available in two Shared Socio-Economic Pathways (SSP2 and SSP5). It is available via Google Earth Engine which makes it subsettable on the server side and the download files relatively lightweight.
We start by reading the config and initializing the Google Earth Engine access again.
Show code cell source
import warnings
warnings.filterwarnings("ignore", category=UserWarning) # Suppress Deprecation Warnings
import configparser
import ast
import geopandas as gpd
import ee
import geemap
import numpy as np
# read local config.ini file
config = configparser.ConfigParser()
config.read('config.ini')
# get paths from config.ini
dir_output = config['FILE_SETTINGS']['DIR_OUTPUT']
dir_figures = config['FILE_SETTINGS']['DIR_FIGURES']
output_gpkg = dir_output + config['FILE_SETTINGS']['GPKG_NAME']
zip_output = config['CONFIG']['ZIP_OUTPUT']
# get style for matplotlib plots
# plt_style = ast.literal_eval(config['CONFIG']['PLOT_STYLE'])
# set the file format for storage
compact_files = config.getboolean('CONFIG','COMPACT_FILES')
# read cloud-project
cloud_project = config['CONFIG']['CLOUD_PROJECT']
# initialize GEE
try:
ee.Initialize(project=cloud_project)
except Exception as e:
ee.Authenticate()
ee.Initialize(project=cloud_project)
Now we can send the catchment outline to GEE to use it as target polygon for aggregation.
Show code cell source
# load catchment outline as target polygon
catchment_new = gpd.read_file(output_gpkg, layer='catchment_new')
catchment = geemap.geopandas_to_ee(catchment_new)
print("Catchment outline converted to GEE polygon.")
# name target subdirectory to be created
cmip_dir = dir_output + 'cmip6/'
Catchment outline converted to GEE polygon.
Select, aggregate, and download downscaled CMIP6 data#
We have designed a class called CMIPDownloader that does everything promised in the heading in one go. The buildFeature() function requests daily catchment wide averages of all available CMIP6 models for individual years. All requested years are stored in an ee.ImageCollection by thegetResult() function. To provide the best basis for bias adjustment, a large overlap of reanalysis and scenario data is recommended. By default, the CMIPDownloader class requests everything between the earliest available date from ERA5 (1979) and the latest available date from CMIP6 (2100). The download() function then starts a given number of parallel requests, each downloading a single year and saving it as a CSV file.
We can simply specify a target location and start the download for both variables individually. If you are in a binder or only have few CPUs available, choose a moderate number of requests to avoid “hickups”. The download time depends on the number of parallel processes, the traffic on the GEE servers and other mysterious factors. If you run this notebook in a binder, it usually doesn’t take more than 5 minutes for both downloads to finish.
Show code cell source
from tools.geetools import CMIPDownloader
downloader_t = CMIPDownloader(var='tas', starty=1979, endy=2100, shape=catchment, processes=30, dir=cmip_dir)
downloader_t.download()
downloader_p = CMIPDownloader(var='pr', starty=1979, endy=2100, shape=catchment, processes=30, dir=cmip_dir)
downloader_p.download()
Initiating download request for NEX-GDDP-CMIP6 data from 1979 to 2100.
Downloading CMIP6 data for variable 'tas': 100%|██████████| 122/122 [01:38<00:00, 1.24it/s]
All downloads complete.
Initiating download request for NEX-GDDP-CMIP6 data from 1979 to 2100.
Downloading CMIP6 data for variable 'pr': 100%|██████████| 122/122 [01:27<00:00, 1.39it/s]
All downloads complete.
We have now downloaded individual files for each year and variable and stored them in output/cmip_dir. To use them as model forcing data, they need to be processed.
Processing CMIP6 data#
The corresponding CMIPProcessor class will read all downloaded CSV files and concatenate them into a single file per scenario. It also checks for consistency and drops models that are not available for individual years or scenarios. It processes variables individually and returns a single dataframe for each of the two scenarios from 1979 to 2100.
Show code cell source
from tools.geetools import CMIPProcessor
cmip_dir = dir_output + 'cmip6/'
processor_t = CMIPProcessor(file_dir=cmip_dir, var='tas')
ssp2_tas_raw, ssp5_tas_raw = processor_t.get_results()
processor_p = CMIPProcessor(file_dir=cmip_dir, var='pr')
ssp2_pr_raw, ssp5_pr_raw = processor_p.get_results()
Let’s have a look. We can see that our scenario dataset now contains a fairly large number of CMIP6 models in alphabetical order.
Show code cell source
print(ssp2_tas_raw.info())
<class 'pandas.core.frame.DataFrame'>
DatetimeIndex: 44560 entries, 1979-01-01 to 2100-12-31
Data columns (total 32 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ACCESS-CM2 44560 non-null float64
1 ACCESS-ESM1-5 44560 non-null float64
2 BCC-CSM2-MR 44560 non-null float64
3 CESM2 44560 non-null float64
4 CESM2-WACCM 44560 non-null float64
5 CMCC-CM2-SR5 44560 non-null float64
6 CMCC-ESM2 44560 non-null float64
7 CNRM-CM6-1 44560 non-null float64
8 CNRM-ESM2-1 44560 non-null float64
9 CanESM5 44560 non-null float64
10 EC-Earth3 44560 non-null float64
11 EC-Earth3-Veg-LR 44560 non-null float64
12 FGOALS-g3 44560 non-null float64
13 GFDL-CM4_gr1 44560 non-null float64
14 GFDL-CM4_gr2 44560 non-null float64
15 GFDL-ESM4 44560 non-null float64
16 GISS-E2-1-G 44560 non-null float64
17 HadGEM3-GC31-LL 44560 non-null float64
18 INM-CM4-8 44560 non-null float64
19 INM-CM5-0 44560 non-null float64
20 IPSL-CM6A-LR 44560 non-null float64
21 KACE-1-0-G 44560 non-null float64
22 KIOST-ESM 44560 non-null float64
23 MIROC-ES2L 44560 non-null float64
24 MIROC6 44560 non-null float64
25 MPI-ESM1-2-HR 44560 non-null float64
26 MPI-ESM1-2-LR 44560 non-null float64
27 MRI-ESM2-0 44560 non-null float64
28 NESM3 44560 non-null float64
29 NorESM2-MM 44560 non-null float64
30 TaiESM1 44560 non-null float64
31 UKESM1-0-LL 44560 non-null float64
dtypes: float64(32)
memory usage: 11.2 MB
None
If we want to check which models failed the consistency check of the CMIPProcessor we can use its dropped_models attribute.
Show code cell source
print('Models that failed the consistency checks:\n')
print(processor_t.dropped_models)
Models that failed the consistency checks:
['HadGEM3-GC31-MM', 'IITM-ESM']
Bias adjustment using reananlysis data#
Due to the coarse resolution of global climate models (GCMs) and the extensive correction of reanalysis data there is substantial bias between the two datasets. To force a glacio-hydrological model calibrated on reanalysis data with climate scenarios this bias needs to be adressed. We will use a method developed by Switanek et.al. (2017) called Scaled Distribution Mapping (SDM) to correct for bias while preserving trends and the likelihood of meteorological events in the raw GCM data. The method has been implemented in the bias_correction Python library by Pankaj Kumar. As suggested by the authors we will apply the bias adjustment to discrete blocks of data individually. The adjust_bias() function loops over all models and adjusts them to the reanalysis data in the overlap period (1979 to 2024).
The function is applied separately to each variable and scenario. The bias_adjustment library provides a normal and a gamma distribution as a basis for the SDM. As the distribution of the ERA5 Land precipitation data is actually closer to a normal distribution with a cut-off of 0 mm, we use the normal_mapping method for both variables.
The function is applied separately to each variable and scenario. The bias_adjustment library provides a normal and a gamma distribution as a basis for the SDM. As the distribution of the ERA5 Land precipitation data is actually closer to a normal distribution with a cut-off of 0 mm, we use the normal_mapping method for both variables.
Show code cell source
from tools.helpers import adjust_bias
era5_file = dir_output + 'ERA5L.csv'
print("SSP2 Temperature:")
ssp2_tas = adjust_bias(predictand=ssp2_tas_raw, predictor=era5_file)
print("SSP5 Temperature:")
ssp5_tas = adjust_bias(predictand=ssp5_tas_raw, predictor=era5_file)
print("SSP2 Precipitation:")
ssp2_pr = adjust_bias(predictand=ssp2_pr_raw, predictor=era5_file)
print("SSP5 Precipitation:")
ssp5_pr = adjust_bias(predictand=ssp5_pr_raw, predictor=era5_file)
SSP2 Temperature:
Bias Correction: 100%|██████████| 12/12 [00:06<00:00, 1.87it/s]
SSP5 Temperature:
Bias Correction: 100%|██████████| 12/12 [00:04<00:00, 2.52it/s]
SSP2 Precipitation:
Bias Correction: 100%|██████████| 12/12 [00:04<00:00, 2.71it/s]
SSP5 Precipitation:
Bias Correction: 100%|██████████| 12/12 [00:04<00:00, 2.61it/s]
The result is a comprehensive dataset of several models over 122 years in two versions (pre- and post-adjustment) for every variable. To see what’s in the data and what happened during bias adjustment we need an overview.
Visualization#
The first plot will contain simple timeseries. The first function cmip_plot() resamples the data so a given frequency and creates a single plot. cmip_plot_combined() arranges multiple plots for both scenarios before and after bias adjustment.
Time series#
First, we store our raw and adjusted data in dictionaries. By default, the data is smoothed with a 10-year moving average (smooting_window=10). Precipitation data is aggregated to annual totals (agg_level='annual'). You can customise this by specifying the appropriate arguments.
Show code cell source
from tools.helpers import read_era5l
from tools.plots import cmip_plot_combined
# store CMIP6 data in dictionaries
ssp_tas_dict = {'SSP2_raw': ssp2_tas_raw, 'SSP2_adjusted': ssp2_tas, 'SSP5_raw': ssp5_tas_raw, 'SSP5_adjusted': ssp5_tas}
ssp_pr_dict = {'SSP2_raw': ssp2_pr_raw, 'SSP2_adjusted': ssp2_pr, 'SSP5_raw': ssp5_pr_raw, 'SSP5_adjusted': ssp5_pr}
era5 = read_era5l(era5_file)
cmip_plot_combined(data=ssp_tas_dict, target=era5, target_label='ERA5-Land', smooth_window=10, show=True, fig_path=f"{dir_figures}NB3_CMIP6_Temp.png")
cmip_plot_combined(data=ssp_pr_dict, target=era5, precip=True, target_label='ERA5-Land', agg_level='annual', smooth_window=10, show=True, fig_path=f"{dir_figures}NB3_CMIP6_Prec.png")
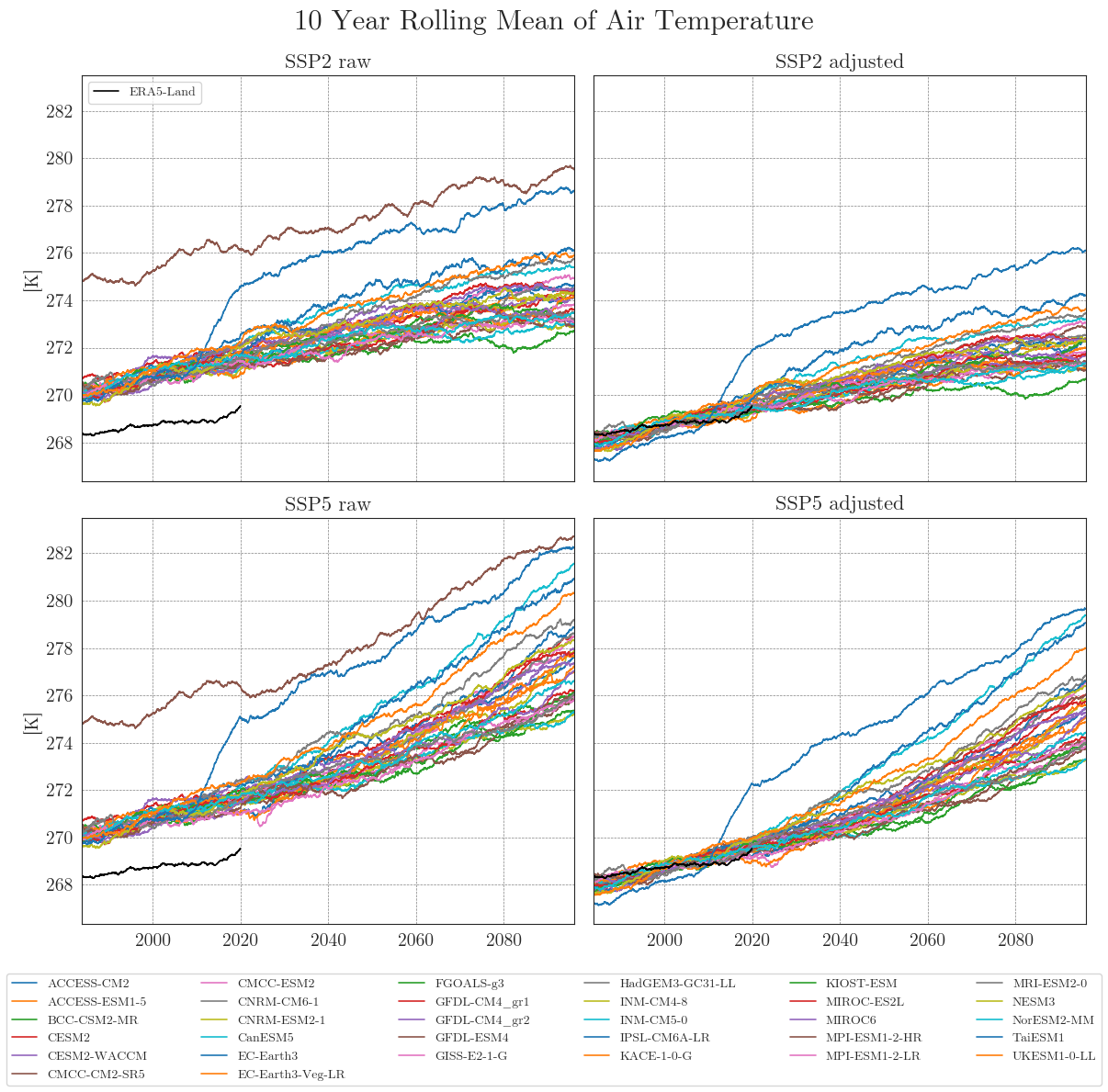
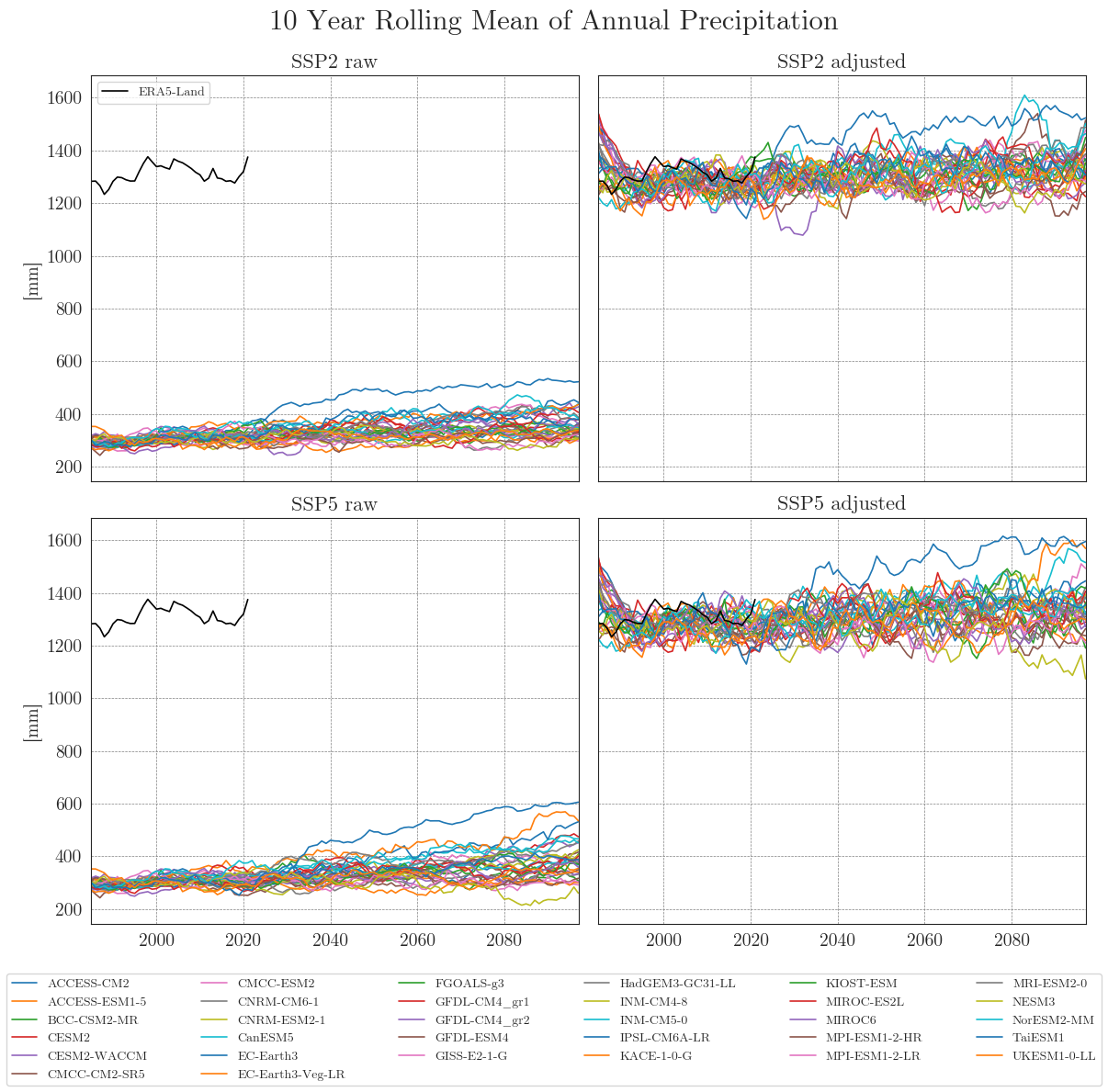
Apparently, some models have striking curves indicating unrealistic outliers or sudden jumps in the data. To clearly identify faulty time series, one option is to choose a qualitative approach by identifying the models using an interactive plotly plot. Here we can zoom and select/deselect curves as we like, to identify model names.
Show code cell source
import plotly.express as px
fig = px.line(ssp5_tas_raw.resample('10YE', closed='left', label='left').mean())
fig.show()
Violin plots#
To look at it from a different perpective we can also have a look at the individual distributions of all models. A nice way to cover several aspects at once is to use seaborne violinplots.
First we have to rearrange our input dictionaries a little bit. For comparison the vplots() function will arrange the plots in a similar grid as in the figures above.
Show code cell source
from tools.helpers import dict_filter
from tools.plots import vplots
tas_raw = dict_filter(ssp_tas_dict, 'raw')
tas_adjusted = dict_filter(ssp_tas_dict, 'adjusted')
pr_raw = dict_filter(ssp_pr_dict, 'raw')
pr_adjusted = dict_filter(ssp_pr_dict, 'adjusted')
vplots(tas_raw, tas_adjusted, era5, target_label='ERA5-Land', show=True, fig_path=f"{dir_figures}NB3_vplot_Temp.png")
vplots(pr_raw, pr_adjusted, era5, target_label='ERA5-Land', precip=True, show=True, fig_path=f"{dir_figures}NB3_vplot_Prec.png")
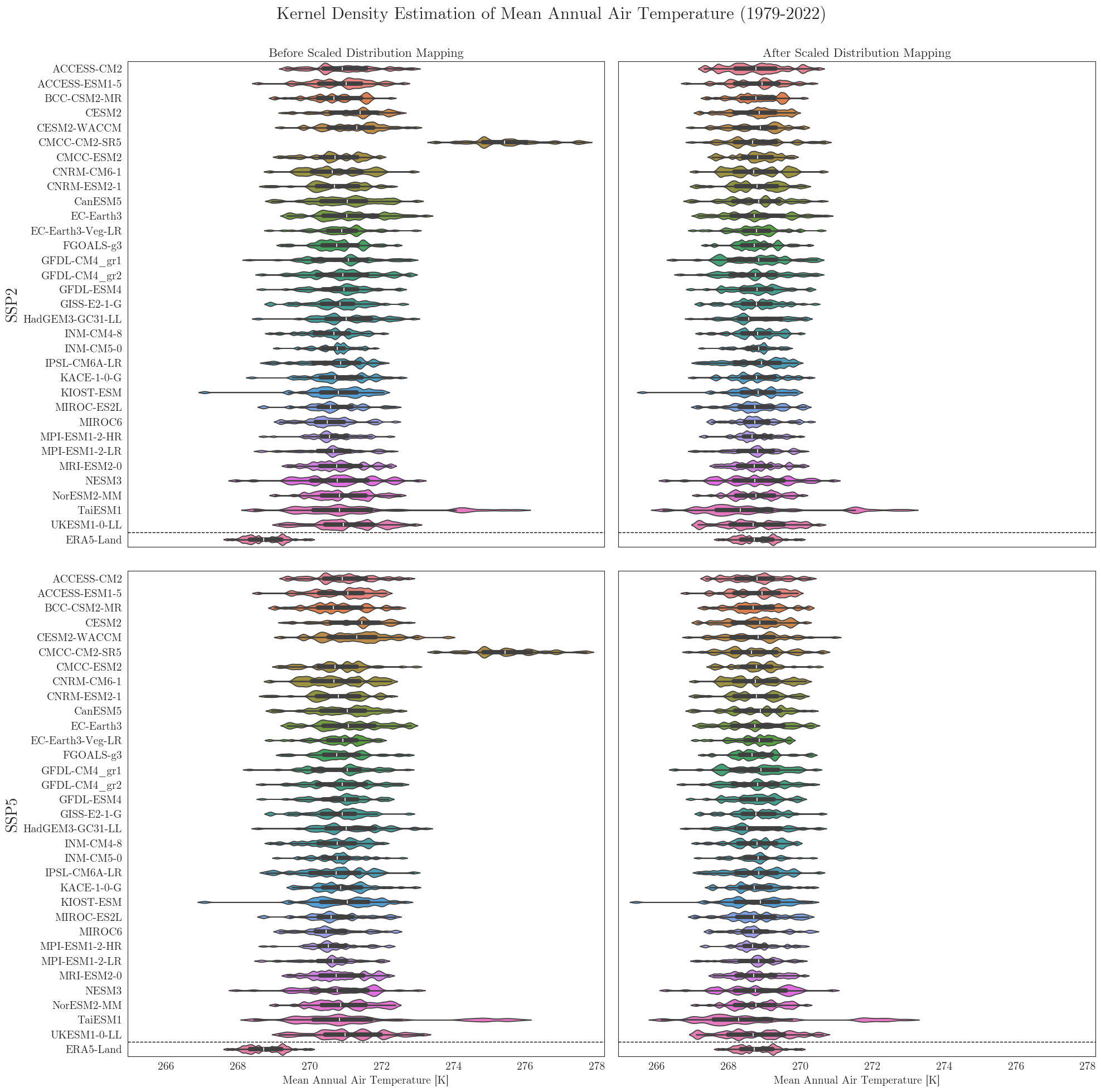
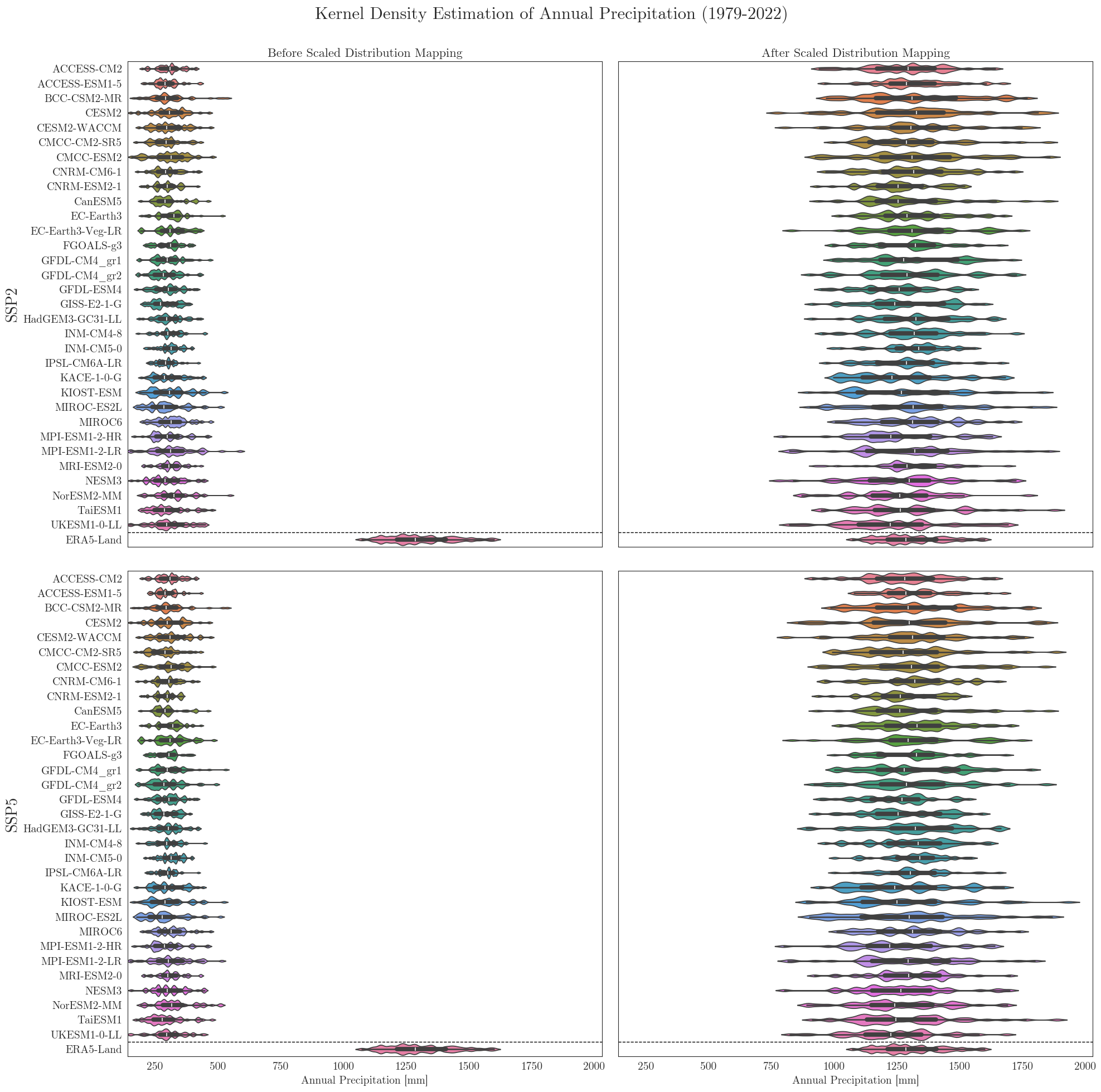
Data filters#
Since we have a large number of models and some problems may be difficult to identify in a plot, we can use some standard filters combined in the DataFilter class. By default it filters models that contain …
… outliers deviating more than 3 standard deviations from the mean (zscore_threshold) and/or …
… increases/decreases of more than 5 units in a single timestep (jump_threshold).
The functions can be applied separately (check_outliers or check_jumps) or together (filter_all). All three return a list of model names.
Here, we also use the resampling_rate argument to resample the data to annual means ('YE') before running the checks.
Show code cell source
from tools.helpers import DataFilter
filter = DataFilter(ssp2_tas_raw, zscore_threshold=3, jump_threshold=5, resampling_rate='YE')
print('Models with temperature outliers: ' + str(filter.outliers))
print('Models with temperature jumps: ' + str(filter.jumps))
print('Models with either one or both: ' + str(filter.filtered_models))
Models with temperature outliers: ['KIOST-ESM']
Models with temperature jumps: []
Models with either one or both: ['KIOST-ESM']
The identified columns can then be removed from the CMIP6 ensemble dataset using another helper function. We run the drop_model() function on the dictionaries of all variables and run cmip_plot_combined() again to check the result.
Show code cell source
from tools.helpers import drop_model
ssp_tas_dict = drop_model(filter.filtered_models, ssp_tas_dict)
ssp_pr_dict = drop_model(filter.filtered_models, ssp_pr_dict)
cmip_plot_combined(data=ssp_tas_dict, target=era5, target_label='ERA5-Land', show=True, fig_path=f"{dir_figures}NB3_CMIP6_Temp_filtered.png")
cmip_plot_combined(data=ssp_pr_dict, target=era5, precip=True, target_label='ERA5-Land', show=True, agg_level='annual', smooth_window=10, fig_path=f"{dir_figures}NB3_CMIP6_Prec_filtered.png")
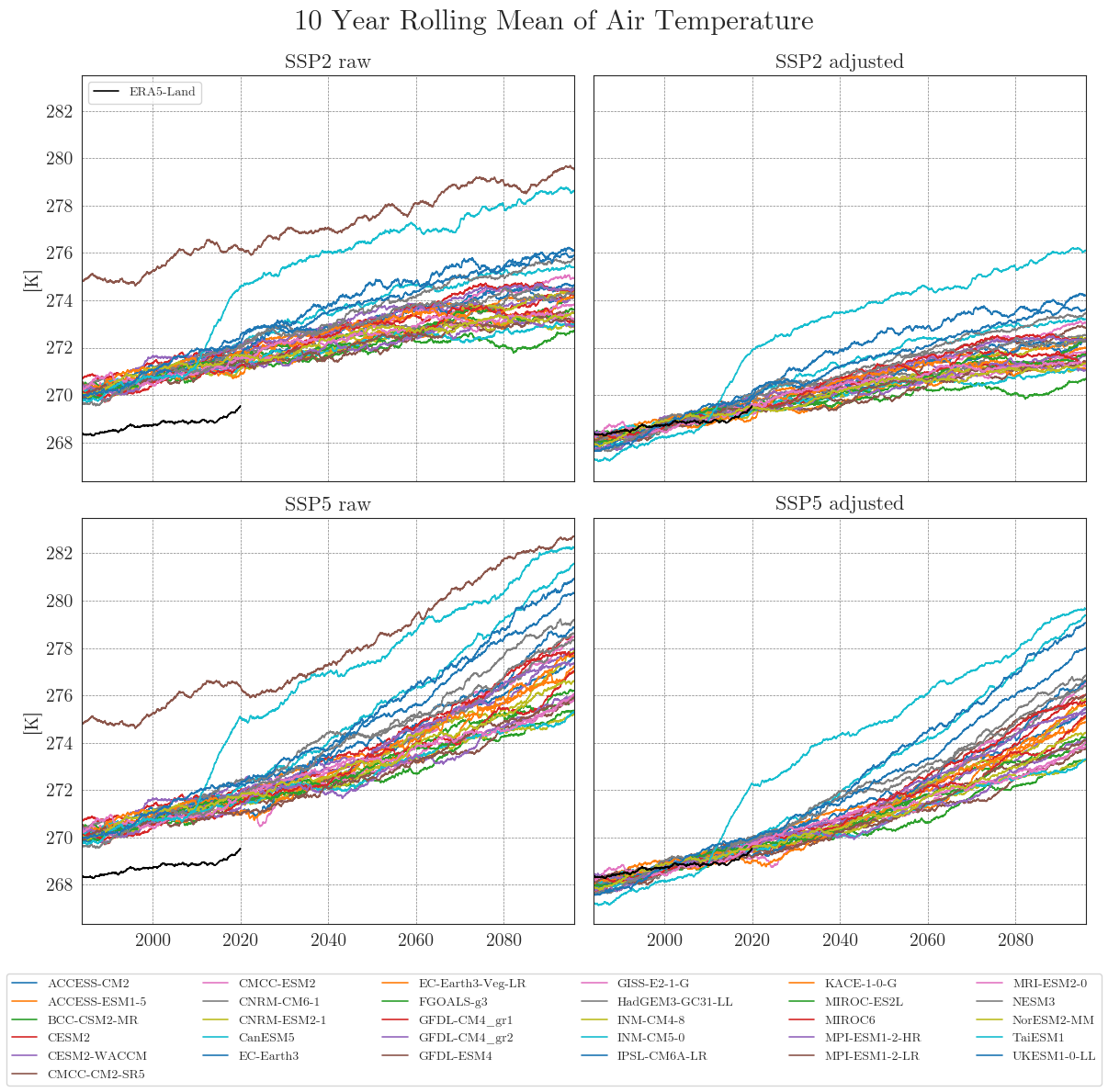
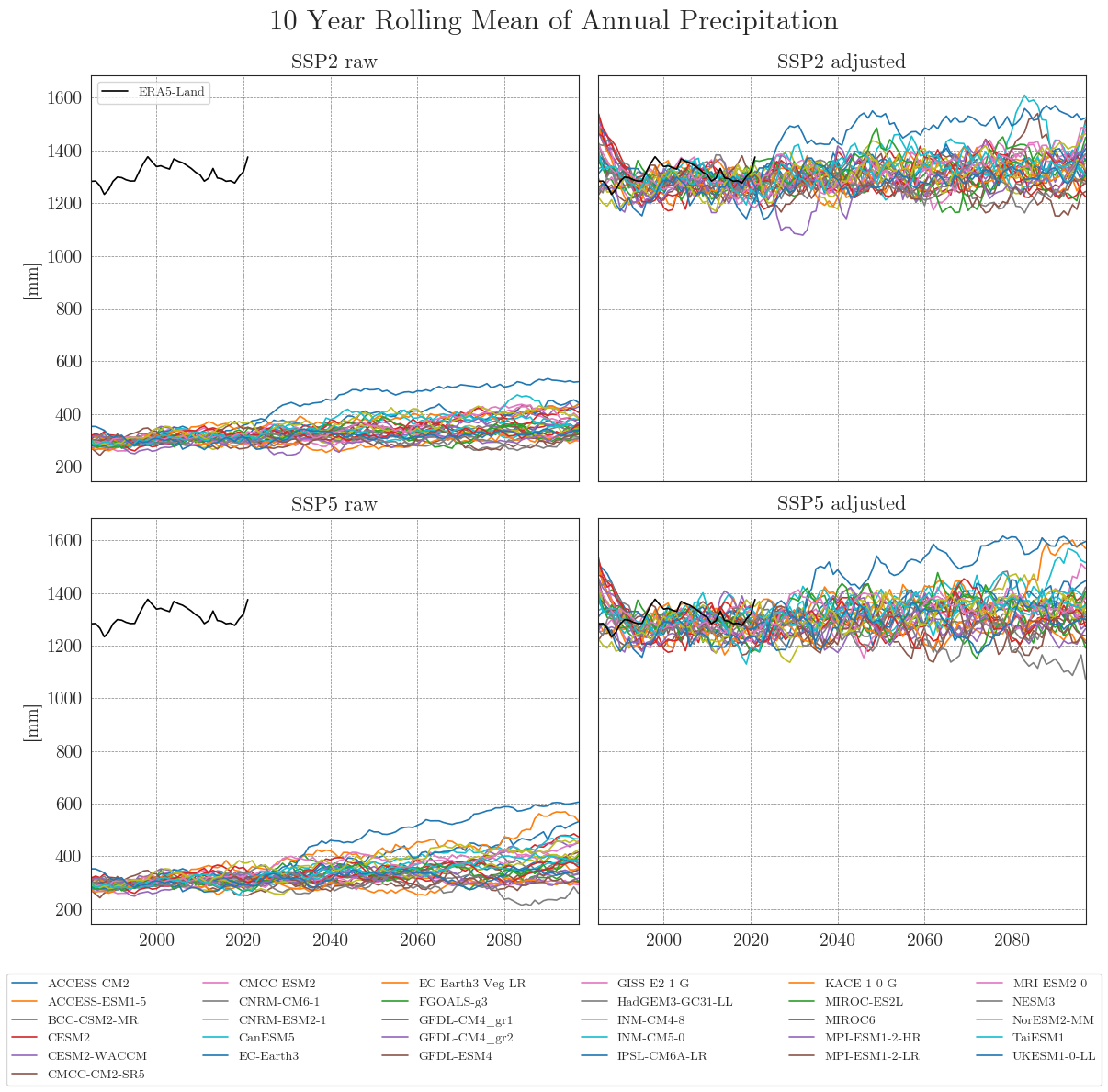
Ensemble mean plots#
Now that we no longer need to focus on individual models, we can reduce the number of lines by plotting only the ensemble means with a 90% confidence interval. With fewer lines in the plot, we can also reduce the resampling frequency and display the annual means.
Show code cell source
from tools.plots import cmip_plot_ensemble
cmip_plot_ensemble(ssp_tas_dict, era5['temp'], intv_mean='YE', fig_path=f'{dir_figures}NB3_CMIP6_Ensemble_Temp.png')
cmip_plot_ensemble(ssp_pr_dict, era5['prec'], precip=True, intv_sum='ME', intv_mean='YE', fig_path=f'{dir_figures}NB3_CMIP6_Ensemble_Prec.png')
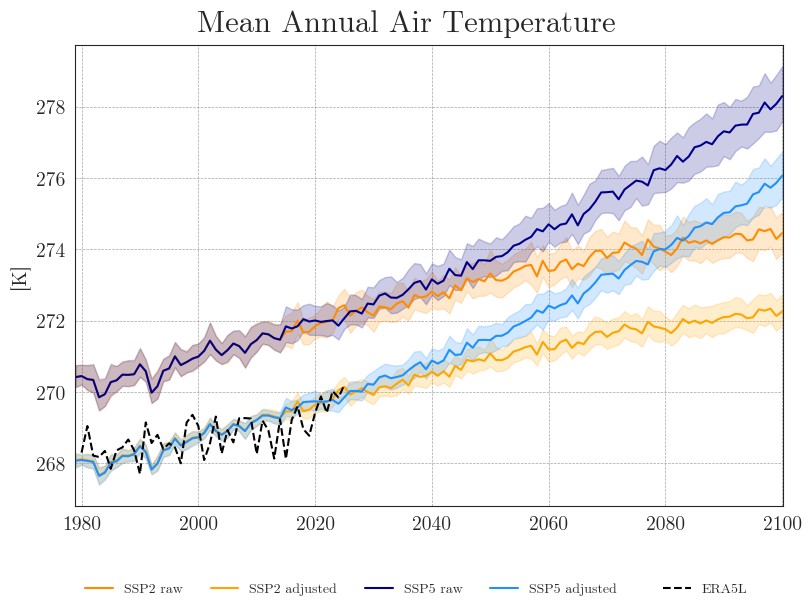
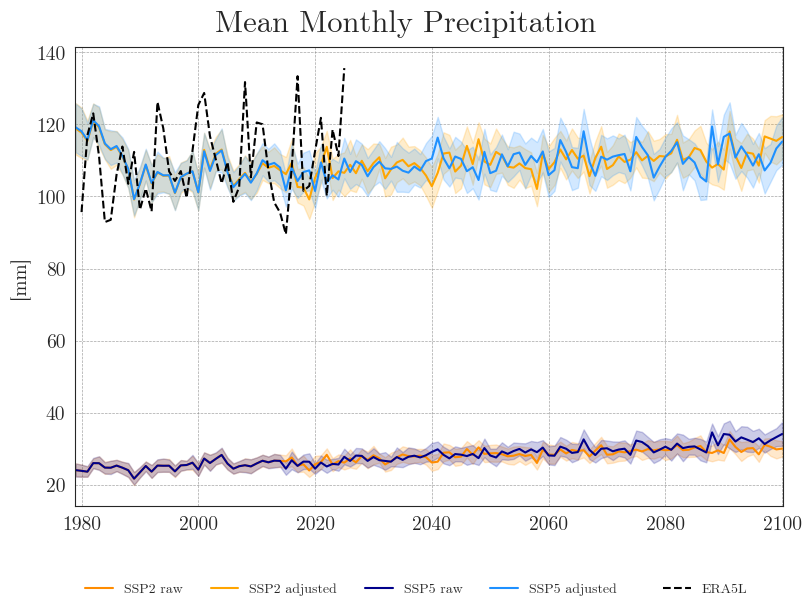
We can see that the SDM adjusts the range and mean of the target data while preserving the distribution and trend of the original data. However, the inter-model variance is slightly reduced for temperature and notably increased for precipitation.
Last but not least, we will have a closer look at the performance of the bias adjustment. To do that, we will create probability plots for all models comparing original, target, and adjusted data with each other and a standard normal distribution. The prob_plot function creates such a plot for an individual model and scenario. The pp_matrix function loops the prob_plot function over all models in a DataFrame and arranges them in a matrix.
First we’ll have a look at the temperature.
Show code cell source
from tools.plots import pp_matrix
pp_matrix(ssp2_tas_raw, era5['temp'], ssp2_tas, scenario='SSP2', show=True, fig_path=f'{dir_figures}NB3_CMIP6_SSP2_probability_Temp.png')
pp_matrix(ssp5_tas_raw, era5['temp'], ssp5_tas, scenario='SSP5', show=True, fig_path=f'{dir_figures}NB3_CMIP6_SSP5_probability_Temp.png')
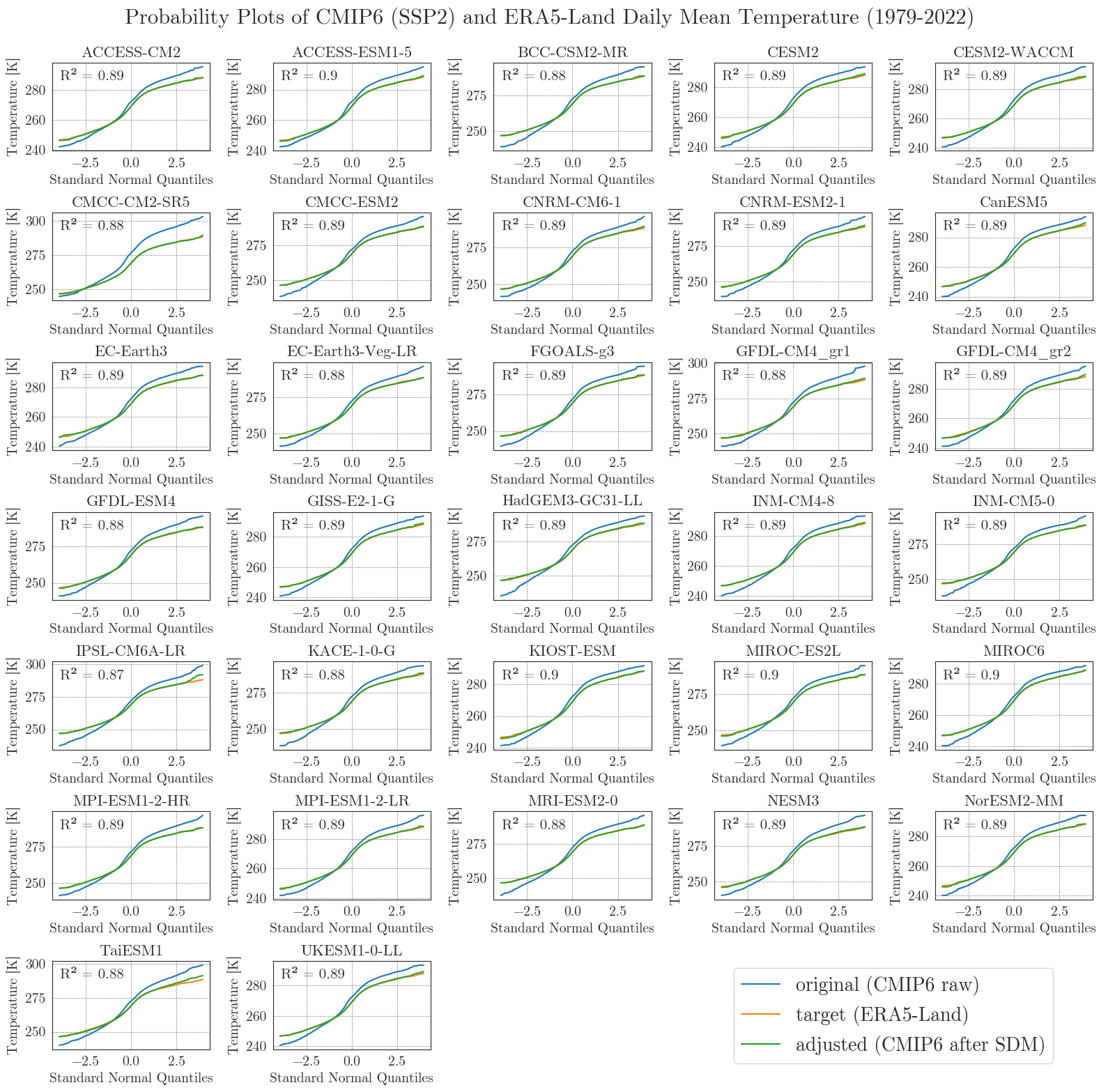
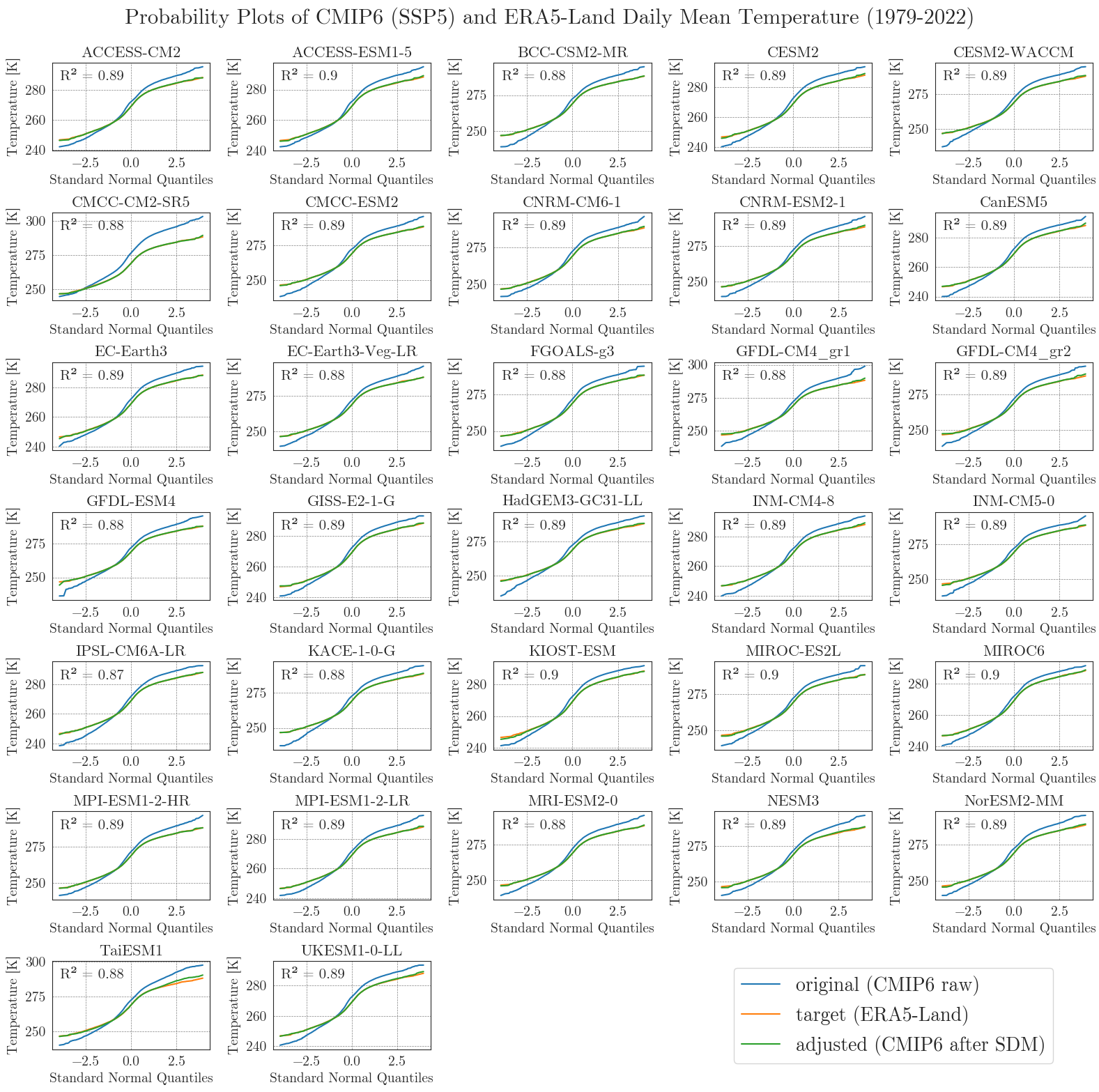
We can see that the SDM worked very well for the temperature data, with high agreement between the target and adjusted data.
Now, let’s examine the probability curves for precipitation. Since the precipitation data is bounded at 0 mm and most days have small values greater than 0 mm, we resample the data as monthly sums to get an idea of the overall performance.
Show code cell source
pp_matrix(ssp2_pr_raw, era5['prec'], ssp2_pr, precip=True, scenario='SSP2', show=True, fig_path=f'{dir_figures}NB3_CMIP6_SSP2_probability_Prec.png')
pp_matrix(ssp5_pr_raw, era5['prec'], ssp5_pr, precip=True, scenario='SSP5', show=True, fig_path=f'{dir_figures}NB3_CMIP6_SSP5_probability_Prec.png')
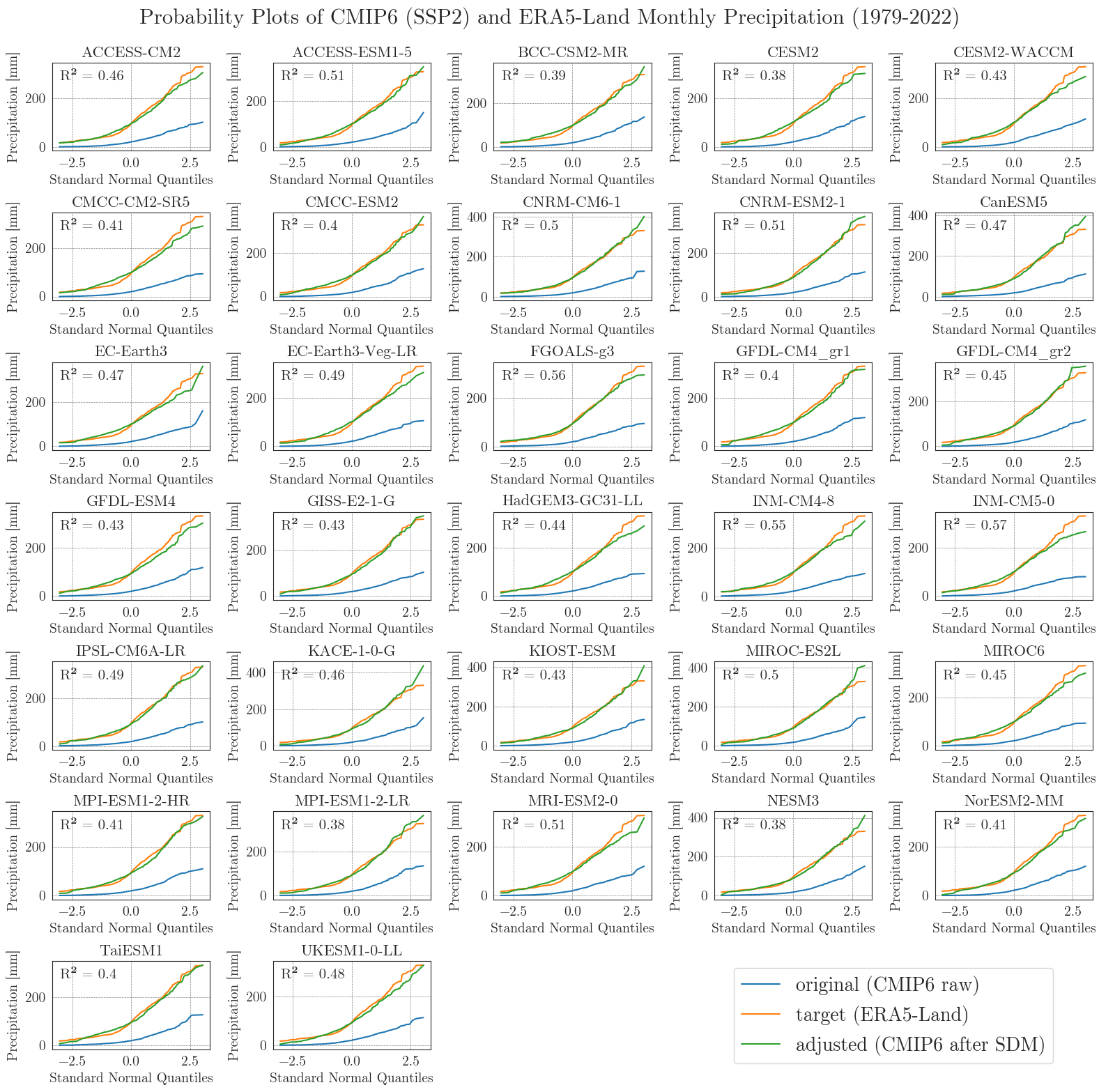
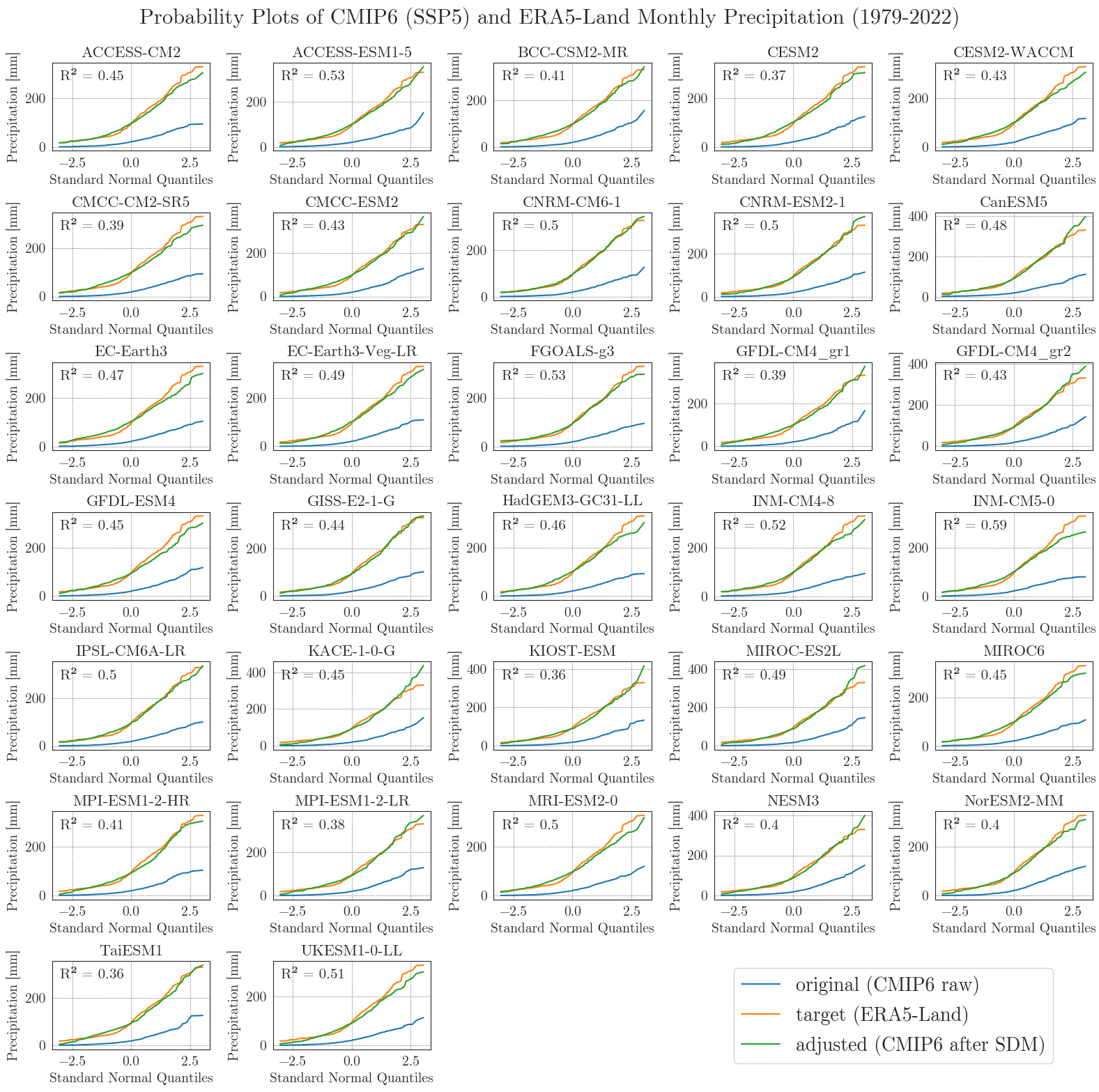
Considering the complexity and heterogeneity of precipitation data, the performance of the SDM is acceptable. Although the fitted data of most models differ from the target data at low and very high values, the general distribution of monthly precipitation is accurately represented.
Write CMIP6 data to file#
After a thorough review of the climate scenario data, we can write the final selection to files to be used in the next notebook. We want to use reanalysis data for the MATILDA model wherever possible and only use CMIP6 data for future projections. Therefore, we need to replace all of the data from our calibration period with ERA5-Land data.
Show code cell source
from tools.helpers import replace_values
era5l = read_era5l(era5_file)
ssp2_tas = ssp_tas_dict['SSP2_adjusted'].copy()
ssp5_tas = ssp_tas_dict['SSP5_adjusted'].copy()
ssp2_pr = ssp_pr_dict['SSP2_adjusted'].copy()
ssp5_pr = ssp_pr_dict['SSP5_adjusted'].copy()
ssp2_tas = replace_values(ssp2_tas, era5l, 'temp')
ssp5_tas = replace_values(ssp5_tas, era5l, 'temp')
ssp2_pr = replace_values(ssp2_pr, era5l, 'prec')
ssp5_pr = replace_values(ssp5_pr, era5l, 'prec')
Since the whole ensemble results in relatively large files, we store the dictionaries in binary format. While these are not human-readable, they are compact and fast to read and write.
pickle files are fast to read and write, but take up more disk space (COMPACT_FILES = False). You can use them on your local machine. parquet files need less disk space but take longer to read and write (COMPACT_FILES = True). They should be your choice in the Binder.Show code cell source
from tools.helpers import dict_to_pickle, dict_to_parquet
import shutil
tas = {'SSP2': ssp2_tas, 'SSP5': ssp5_tas}
pr = {'SSP2': ssp2_pr, 'SSP5': ssp5_pr}
if compact_files:
# For storage efficiency:
dict_to_parquet(tas, cmip_dir + 'adjusted/tas_parquet')
dict_to_parquet(pr, cmip_dir + 'adjusted/pr_parquet')
else:
# For speed:
dict_to_pickle(tas, cmip_dir + 'adjusted/tas.pickle')
dict_to_pickle(pr, cmip_dir + 'adjusted/pr.pickle')
if zip_output:
# refresh `output_download.zip` with data retrieved within this notebook
shutil.make_archive('output_download', 'zip', 'output')
print('Output folder can be download now (file output_download.zip)')
Output folder can be download now (file output_download.zip)
Show code cell source
%reset -f
You can now continue with Notebook 4 to calibrate the MATILDA model.
