Climate forcing data#
Now that we have all the static data, we can focus on the climate variables. In this notebook we will…
…download ERA5 land reanalysis data aggregated to our catchment,
…and determine the reference altitude of the data from the geopotential height.
For data preprocessing and download we will again use the Google Earth Engine (GEE) to offload as much as possible to external servers. ERA5-Land is the latest reanalysis dataset from the European Center for Medium-Range Weather Forecast (ECMWF), available from 1950 to near real-time. The GEE data catalog summarizes…
ERA5-Land is a reanalysis dataset providing a consistent view of the evolution of land variables over several decades at an enhanced resolution compared to ERA5. ERA5-Land has been produced by replaying the land component of the ECMWF ERA5 climate reanalysis. Reanalysis combines model data with observations from across the world into a globally complete and consistent dataset using the laws of physics. Reanalysis produces data that goes several decades back in time, providing an accurate description of the climate of the past.
Source: GEE Data Catalog
To get started we read some settings from the config.ini file again:
cloud project name for the GEE access
input/output folders for data imports and downloads
filenames (DEM, GeoPackage)
include future projections or not
show/hide interactive map in notebooks
Show code cell source
import warnings
warnings.filterwarnings("ignore", category=UserWarning) # Suppress Deprecation Warnings
import pandas as pd
import configparser
import ast
# read local config.ini file
config = configparser.ConfigParser()
config.read('config.ini')
# get file config from config.ini
cloud_project = config['CONFIG']['CLOUD_PROJECT']
dir_input = config['FILE_SETTINGS']['DIR_INPUT']
dir_output = config['FILE_SETTINGS']['DIR_OUTPUT']
dir_figures = config['FILE_SETTINGS']['DIR_FIGURES']
output_gpkg = dir_output + config['FILE_SETTINGS']['GPKG_NAME']
scenarios = config.getboolean('CONFIG', 'PROJECTIONS')
show_map = config.getboolean('CONFIG','SHOW_MAP')
zip_output = config['CONFIG']['ZIP_OUTPUT']
# get style for matplotlib plots
plt_style = ast.literal_eval(config['CONFIG']['PLOT_STYLE'])
…and initialize the GEE API.
Show code cell source
import ee
try:
ee.Initialize(project=cloud_project)
except Exception as e:
ee.Authenticate()
ee.Initialize(project=cloud_project)
We can now load the catchment outline from the previous notebook and convert it to a ee.FeatureCollection to use it in GEE.
Show code cell source
import geopandas as gpd
import geemap
catchment_new = gpd.read_file(output_gpkg, layer='catchment_new')
catchment = geemap.geopandas_to_ee(catchment_new)
Set the date range#
If you are only interested in modeling the past, set PROJECTIONS=False in the config.ini to only download reanalysis data for your defined modeling period. Otherwise, all available historical data (since 1979) is downloaded to provide the best possible basis for bias adjustment of the climate scenario data.
Show code cell source
if scenarios == True:
date_range = ['1979-01-01', '2025-01-01']
else:
date_range = ast.literal_eval(config['CONFIG']['DATE_RANGE'])
print(f'The selected date range is {date_range[0]} to {date_range[1]}')
The selected date range is 1979-01-01 to 2025-01-01
ERA5L Geopotential height#
The reference surface elevation of ERA5-Land grid cells cannot be obtained directly, but must be calculated from the geopotential.
This parameter is the gravitational potential energy of a unit mass, at a particular location, relative to mean sea level. It is also the amount of work that would have to be done, against the force of gravity, to lift a unit mass to that location from mean sea level.
The geopotential height can be calculated by dividing the geopotential by the Earth’s gravitational acceleration, g (=9.80665 m s-2). The geopotential height plays an important role in synoptic meteorology (analysis of weather patterns). Charts of geopotential height plotted at constant pressure levels (e.g., 300, 500 or 850 hPa) can be used to identify weather systems such as cyclones, anticyclones, troughs and ridges.
At the surface of the Earth, this parameter shows the variations in geopotential (height) of the surface, and is often referred to as the orography.
Source: ECMWF Parameter Database
Since the ERA5 geopotential height is not available in the GEE Data Catalog, we downloaded it using the ECMWF Copernicus Data Store (CDS) API, converted it to .ncdf format, and reuploaded it. Therefore, the file has to be accessed in a similar way to the ice thickness data in Notebook 1.
Show code cell source
from resourcespace import ResourceSpace
# use guest credentials to access media server
api_base_url = config['MEDIA_SERVER']['api_base_url']
private_key = config['MEDIA_SERVER']['private_key']
user = config['MEDIA_SERVER']['user']
myrepository = ResourceSpace(api_base_url, user, private_key)
# get resource IDs for each .zip file
refs_era5l = pd.DataFrame(myrepository.get_collection_resources(128))[['ref', 'file_size', 'file_extension', 'field8']]
ref_geopot = refs_era5l.loc[refs_era5l['field8'] == 'ERA5_land_Z_geopotential']
print("Dataset file and reference on media server:\n")
display(ref_geopot)
Dataset file and reference on media server:
| ref | file_size | file_extension | field8 | |
|---|---|---|---|---|
| 0 | 27215 | 8062521 | zip | ERA5_land_Z_geopotential |
The .ncdf file is then unzipped and loaded as xarray dataset for further processing.
Show code cell source
from zipfile import ZipFile
import io
import xarray as xr
content = myrepository.get_resource_file(ref_geopot.at[0,'ref'])
with ZipFile(io.BytesIO(content), 'r') as zipObj:
# Get a list of all archived file names from the zip
filename = zipObj.namelist()[0]
print(f'Reading file "{filename}"...')
file_bytes = zipObj.read(filename)
# Open the file-like object as an xarray dataset
ds = xr.open_dataset(io.BytesIO(file_bytes))
print(f'Dataset contains {ds.z.attrs["long_name"]} in {ds.z.attrs["units"]} as variable \'{ds.z.attrs["GRIB_cfVarName"]}\'')
Reading file "ERA5_land_Z_geopotential.nc"...
Dataset contains Geopotential in m**2 s**-2 as variable 'z'
The original dataset covers the entire globe, so we crop it to the catchment area plus a 1° buffer zone.
Show code cell source
# get catchment bounding box with buffer
bounds = catchment_new.total_bounds
min_lon = bounds[0] - 1
min_lat = bounds[1] - 1
max_lon = bounds[2] + 1
max_lat = bounds[3] + 1
cropped_ds = ds.sel(lat=slice(min_lat,max_lat), lon=slice(min_lon,max_lon))
print(f"xr.Dataset cropped to bbox[{round(min_lon, 2)}, {round(min_lat, 2)}, {round(max_lon, 2)}, {round(max_lat)}]")
xr.Dataset cropped to bbox[77.06, 41.05, 79.33, 43]
To load xarray data into GEE a little workaround is needed. Credits to Oliver Lopez for this solution.
Show code cell source
# function to load nc file into GEE
import numpy as np
def netcdf_to_ee(ds):
data = ds['z']
lon_data = np.round(data['lon'], 3)
lat_data = np.round(data['lat'], 3)
dim_lon = np.unique(np.ediff1d(lon_data).round(3))
dim_lat = np.unique(np.ediff1d(lat_data).round(3))
if (len(dim_lon) != 1) or (len(dim_lat) != 1):
print("The netCDF file is not a regular longitude/latitude grid")
print("Converting xarray to numpy array...")
data_np = np.array(data)
data_np = np.transpose(data_np)
# Figure out if we need to roll the data or not
# (see https://github.com/giswqs/geemap/issues/285#issuecomment-791385176)
if np.max(lon_data) > 180:
data_np = np.roll(data_np, 180, axis=0)
west_lon = lon_data[0] - 180
else:
west_lon = lon_data[0]
print("Saving data extent and origin...")
transform = [dim_lon[0], 0, float(west_lon) - dim_lon[0]/2, 0, dim_lat[0], float(lat_data[0]) - dim_lat[0]/2]
print("Converting numpy array to ee.Array...")
image = geemap.numpy_to_ee(
data_np, "EPSG:4326", transform=transform, band_names='z'
)
print("Done!")
return image, data_np, transform
image, data_np, transform = netcdf_to_ee(cropped_ds)
Converting xarray to numpy array...
Saving data extent and origin...
Converting numpy array to ee.Array...
Done!
If mapping is enabled in the config.ini you can now display the geopotential data in the GEE map.
Show code cell source
import geemap.colormaps as cm
if show_map:
Map = geemap.Map()
# add geopotential as layer
vis_params = {'min': int(data_np.min()), 'max': int(data_np.max()), 'palette': cm.palettes.terrain, 'opacity': 0.8}
Map.addLayer(image, vis_params, "ERA5L geopotential")
# add catchment
Map.addLayer(catchment, {'color': 'darkgrey'}, "Catchment")
Map.centerObject(catchment, zoom=9)
display(Map)
else:
print("Map view disabled in config.ini")
Since our workflow will use a lumped model, we will use area-weighted catchment-wide averages of our forcing data. Thus, we also aggregate the geopotential based on the grid cell fractions in the catchment and convert it to geopotential height in meters above sea level. This represents the reference altitude of our forcing data, just as the elevation of a weather station would.
Show code cell source
# execute reducer
dict = image.reduceRegion(ee.Reducer.mean(),
geometry=catchment,
crs='EPSG:4326',
crsTransform=transform)
# get mean value and print
mean_val = dict.getInfo()['z']
ele_dat = mean_val / 9.80665
print(f'Geopotential mean:\t{mean_val:.2f} m2 s-2\nElevation:\t\t {ele_dat:.2f} m a.s.l.')
Geopotential mean: 32602.75 m2 s-2
Elevation: 3324.56 m a.s.l.
ERA5-Land Temperature and Precipitation Data#
Our model only requires temperature and precipitation as inputs. We will download both time series from the ERA5-Land Daily Aggregated - ECMWF Climate Reanalysis ECMWF/ERA5_LAND/DAILY_RAW dataset in the Google Earth Engine Data Catalog
The asset is a daily aggregate of ECMWF ERA5 Land hourly assets. […] Daily aggregates have been pre-calculated to facilitate many applications requiring easy and fast access to the data.
Source: GEE Data Catalog
On the server side, we simply create an ee.ImageCollection with the desired bands (temperature and precipitation) and date range. To calculate area-weighted aggregates we apply the ee.Reducer function.
Show code cell source
import pandas as pd
import datetime
def setProperty(image):
dict = image.reduceRegion(ee.Reducer.mean(), catchment)
return image.set(dict)
collection = ee.ImageCollection('ECMWF/ERA5_LAND/DAILY_RAW')\
.select('temperature_2m','total_precipitation_sum')\
.filterDate(date_range[0], date_range[1])
withMean = collection.map(setProperty)
We can then aggregate the results into arrays, download them with .getInfo() and store them as dataframe columns. Depending on the selected date range and server traffic this can take up to a few minutes.
Show code cell source
%%time
df = pd.DataFrame()
print("Get timestamps...")
df['ts'] = withMean.aggregate_array('system:time_start').getInfo()
df['dt'] = df['ts'].apply(lambda x: datetime.datetime.fromtimestamp(x / 1000))
print("Get temperature values...")
df['temp'] = withMean.aggregate_array('temperature_2m').getInfo()
df['temp_c'] = df['temp'] - 273.15
print("Get precipitation values...")
df['prec'] = withMean.aggregate_array('total_precipitation_sum').getInfo()
df['prec'] = df['prec'] * 1000
Get timestamps...
Get temperature values...
Get precipitation values...
CPU times: user 270 ms, sys: 200 ms, total: 470 ms
Wall time: 3min 35s
The constructed data frame now looks like this:
Show code cell source
display(df)
| ts | dt | temp | temp_c | prec | |
|---|---|---|---|---|---|
| 0 | 283996800000 | 1979-01-01 01:00:00 | 257.460053 | -15.689947 | 0.027150 |
| 1 | 284083200000 | 1979-01-02 01:00:00 | 256.509735 | -16.640265 | 0.004800 |
| 2 | 284169600000 | 1979-01-03 01:00:00 | 257.944142 | -15.205858 | 0.001599 |
| 3 | 284256000000 | 1979-01-04 01:00:00 | 258.392049 | -14.757951 | 0.281500 |
| 4 | 284342400000 | 1979-01-05 01:00:00 | 258.125576 | -15.024424 | 0.107802 |
| ... | ... | ... | ... | ... | ... |
| 16797 | 1735257600000 | 2024-12-27 01:00:00 | 263.179270 | -9.970730 | 0.026296 |
| 16798 | 1735344000000 | 2024-12-28 01:00:00 | 259.782440 | -13.367560 | 0.000917 |
| 16799 | 1735430400000 | 2024-12-29 01:00:00 | 261.269940 | -11.880060 | 0.005262 |
| 16800 | 1735516800000 | 2024-12-30 01:00:00 | 263.580786 | -9.569214 | 1.161816 |
| 16801 | 1735603200000 | 2024-12-31 01:00:00 | 259.719079 | -13.430921 | 1.057546 |
16802 rows × 5 columns
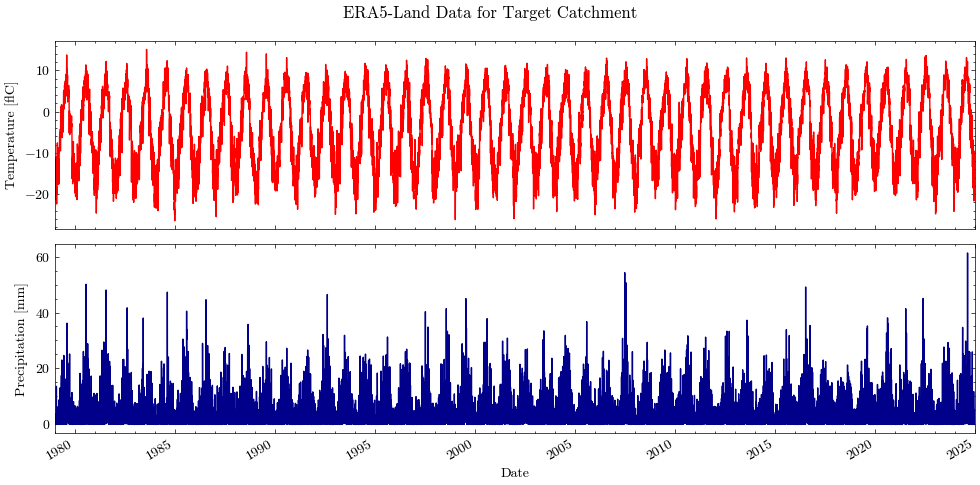
Let’s plot the full time series.
Show code cell source
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
import scienceplots
# set style from config
plt.style.use(plt_style)
axes = df.drop(['ts','temp'],axis=1).plot.line(x='dt', subplots=True, legend=False, figsize=(10,5),
title='ERA5-Land Data for Target Catchment',
color={"temp_c": "red", "prec": "darkblue"})
axes[0].set_ylabel("Temperature [°C]")
axes[1].set_ylabel("Precipitation [mm]")
axes[1].set_xlabel("Date")
axes[1].xaxis.set_minor_locator(mdates.YearLocator())
plt.xlim(date_range)
plt.tight_layout()
plt.savefig(dir_figures+'NB2_ERA5_Temp_Prec.png')
plt.show()
Store data for next steps#
To continue in the workflow, we write the ERA5 data to a .csv file…
Show code cell source
df.to_csv(dir_output + 'ERA5L.csv',header=True,index=False)
…and update the settings.yml file with the reference altitude of the ERA5-Land data (ele_dat) and refresh output_download.zip with newly acquired data.
Show code cell source
from tools.helpers import update_yaml
import shutil
# update settings file
update_yaml(dir_output + 'settings.yml', {'ele_dat': float(ele_dat)})
if zip_output:
# refresh `output_download.zip` with data retrieved within this notebook
shutil.make_archive('output_download', 'zip', 'output')
print('Output folder can be download now (file output_download.zip)')
Data successfully written to YAML at output/settings.yml
Output folder can be download now (file output_download.zip)
Show code cell source
%reset -f
You can now continue with Notebook 3.
