Catchment delineation#
We start our workflow by downloading all the static data we need. In this notebook we will…
…download a Digital Elevation Model (DEM) for hydrologic applications,
…delineate the catchment and determine the catchment area using your reference position (e.g. the location of your gauging station) as the “pouring point”,
…identify all glaciers within the catchment and download the glacier outlines and ice thicknesses,
…create a glacier mass profile based on elevation zones.
First of all, we will read some settings from the config.ini file:
cloud project name for the GEE access
input/output folders for data imports and downloads
filenames (DEM, GeoPackage)
coordinates of the defined “pouring” point (Lat/Long)
chosen DEM from GEE data catalog
show/hide GEE map in notebooks
Show code cell source
import warnings
warnings.filterwarnings("ignore", category=UserWarning) # Suppress Deprecation Warnings
import os
import pandas as pd
import numpy as np
import configparser
import ast
import matplotlib.pyplot as plt
import scienceplots
# read local config.ini file
config = configparser.ConfigParser()
config.read('config.ini')
# get file config from config.ini
cloud_project = config['CONFIG']['CLOUD_PROJECT']
output_folder = config['FILE_SETTINGS']['DIR_OUTPUT']
figures_folder = config['FILE_SETTINGS']['DIR_FIGURES']
filename = output_folder + config['FILE_SETTINGS']['DEM_FILENAME']
output_gpkg = output_folder + config['FILE_SETTINGS']['GPKG_NAME']
zip_output = config['CONFIG']['ZIP_OUTPUT']
# create folder for output figures
os.makedirs(figures_folder, exist_ok=True)
# get used GEE DEM, coords and other settings
dem_config = ast.literal_eval(config['CONFIG']['DEM'])
y, x = ast.literal_eval(config['CONFIG']['COORDS'])
show_map = config.getboolean('CONFIG', 'SHOW_MAP')
# get style for matplotlib plots
plt_style = ast.literal_eval(config['CONFIG']['PLOT_STYLE'])
plt.style.use(plt_style)
# print config data
print(f'Google Cloud Project : {cloud_project}')
print(f'DEM to download: {dem_config[3]}')
print(f'Coordinates of discharge point: Lat {y}, Lon {x}')
Google Cloud Project : huklima
DEM to download: MERIT 30m
Coordinates of discharge point: Lat 42.300264, Lon 78.091444
Now, the Google Earth Engine (GEE) access can be initialized. If this is the first time you run the notebook on this machine, you need to authenticate. When using mybinder.org you need to authenticate every time a new session has been launched. Follow the instructions on screen or see the guide in → Notebook 0.
config.ini
needs to be changed to your own. See the instructions in
Notebook 0, step 5
for details.
Show code cell source
import ee
# initialize GEE at the beginning of session
try:
ee.Initialize(project=cloud_project)
except Exception as e:
ee.Authenticate() # authenticate when using GEE for the first time
ee.Initialize(project=cloud_project)
print("Google Earth Engine Access initialized!")
Google Earth Engine Access initialized!
Start GEE and download DEM#
Once we are set up, we can start working with the data. Let’s start with the base map, if enabled in config.ini. The map can be used to follow the steps as more layers are added throughout the notebook.
Show code cell source
import geemap
if show_map:
Map = geemap.Map()
display(Map)
else:
print("Map view disabled in config.ini")
Now we can download the DEM from the GEE catalog and add it as a new layer to the map. The default is the [MERIT DEM] (https://developers.google.com/earth-engine/datasets/catalog/MERIT_DEM_v1_0_3), but you can use any DEM available in the Google Earth Engine Data Catalog (https://developers.google.com/earth-engine/datasets/catalog) by specifying it in the config.ini file.
Show code cell source
if dem_config[0] == 'Image':
image = ee.Image(dem_config[1]).select(dem_config[2])
elif dem_config[0] == 'ImageCollection':
image = ee.ImageCollection(dem_config[1]).select(dem_config[2]).mosaic()
if show_map:
srtm_vis = {'bands': dem_config[2],
'min': 0,
'max': 6000,
'palette': ['000000', '478FCD', '86C58E', 'AFC35E', '8F7131', 'B78D4F', 'E2B8A6', 'FFFFFF']
}
Map.addLayer(image, srtm_vis, dem_config[3], True, 0.7)
Next, we add the location of our discharge observations to the map and generate a 40km buffer box.
Show code cell source
point = ee.Geometry.Point(x, y)
box = point.buffer(40000).bounds()
if show_map:
Map.addLayer(point, {'color': 'blue'}, 'Discharge Point')
Map.addLayer(box, {'color': 'grey'}, 'Catchment Area', True, 0.7)
Map.centerObject(box, zoom=9)
The gauging location (marker) and the box (polygon/rectangle) can also be added manually to the map above. If features have been drawn, they will overrule the configured discharge point and automatically created box.
Show code cell source
if show_map:
for feature in Map.draw_features:
f_type = feature.getInfo()['geometry']['type']
if f_type == 'Point':
point = feature.geometry()
print("Manually set pouring point will be considered")
elif f_type == 'Polygon':
box = feature.geometry()
print("Manually drawn box will be considered")
Now we can export the DEM as a .tif file for the selected extent to the output folder. Depending on the size of the selected area, this might take a while for processing and downloading.
Show code cell source
import xarray as xr
download_xr = config.getboolean('CONFIG', 'GEE_DOWNLOAD_XR')
if download_xr:
# new method using Xarray (supports larger areas)
try:
print('Get GEE data as Xarray...')
ic = ee.ImageCollection(image)
ds = xr.open_dataset(
ic,
engine='ee',
projection=ic.first().select(0).projection(),
geometry=box
)
print('Prepare Xarray for GeoTiff conversion...')
ds_t = ds.isel(time=0).drop_vars("time").transpose()
ds_t.rio.set_spatial_dims("lon", "lat", inplace=True)
print('Save DEM as GeoTiff...')
ds_t.rio.to_raster(filename)
print('DEM successfully saved at', filename)
except:
print('Error during Xarray routine. Try direct download from GEE...')
geemap.ee_export_image(image, filename=filename, scale=30, region=box, file_per_band=False)
else:
# old method using GEE API to download .tif directly
geemap.ee_export_image(image, filename=filename, scale=30, region=box, file_per_band=False)
Get GEE data as Xarray...
Prepare Xarray for GeoTiff conversion...
Save DEM as GeoTiff...
DEM successfully saved at output/dem_gee.tif
Catchment deliniation#
Based on the downloaded DEM file, we can delineate the watershed using the pysheds library. The result will be a raster and displayed at the end of this section.
The full documentation of the pysheds module can be found here.
Show code cell source
%%time
# GIS packages
from pysheds.grid import Grid
import fiona
# load DEM
DEM_file = filename
grid = Grid.from_raster(DEM_file)
dem = grid.read_raster(DEM_file)
print("DEM loaded.")
DEM loaded.
CPU times: user 1.21 s, sys: 184 ms, total: 1.39 s
Wall time: 1.63 s
Show code cell source
%%time
# Fill depressions in DEM
print("Fill depressions in DEM...")
flooded_dem = grid.fill_depressions(dem)
# Resolve flats in DEM
print("Resolve flats in DEM...")
inflated_dem = grid.resolve_flats(flooded_dem)
# Specify directional mapping
# N NE E SE S SW W NW
dirmap = (64, 128, 1, 2, 4, 8, 16, 32)
# Compute flow directions
print("Compute flow directions...")
fdir = grid.flowdir(inflated_dem, dirmap=dirmap)
# catch = grid.catchment(x=x, y=y, fdir=fdir, dirmap=dirmap, xytype='coordinate')
# Compute accumulation
print("Compute accumulation...")
acc = grid.accumulation(fdir)
# Snap pour point to high accumulation cell
x_snap, y_snap = grid.snap_to_mask(acc > 1000, (x, y))
# Delineate the catchment
print("Delineate the catchment...")
catch = grid.catchment(x=x_snap, y=y_snap, fdir=fdir, xytype='coordinate')
# Clip the DEM to the catchment
print("Clip the DEM to the catchment...")
grid.clip_to(catch)
clipped_catch = grid.view(catch)
print("Processing completed.")
Fill depressions in DEM...
Resolve flats in DEM...
Compute flow directions...
Compute accumulation...
Delineate the catchment...
Clip the DEM to the catchment...
Processing completed.
CPU times: user 1.54 s, sys: 93.9 ms, total: 1.64 s
Wall time: 1.71 s
Now let’s have a look at the catchment area.
Show code cell source
# Define a function to plot the digital elevation model
def plotFigure(data, label, cmap='Blues'):
plt.figure(figsize=(12, 10))
plt.imshow(data, extent=grid.extent, cmap=cmap)
plt.colorbar(label=label)
plt.grid()
demView = grid.view(dem, nodata=np.nan)
plotFigure(demView, 'Elevation in Meters', cmap='terrain')
plt.savefig(figures_folder + 'NB1_DEM_Catchment.png')
plt.show()
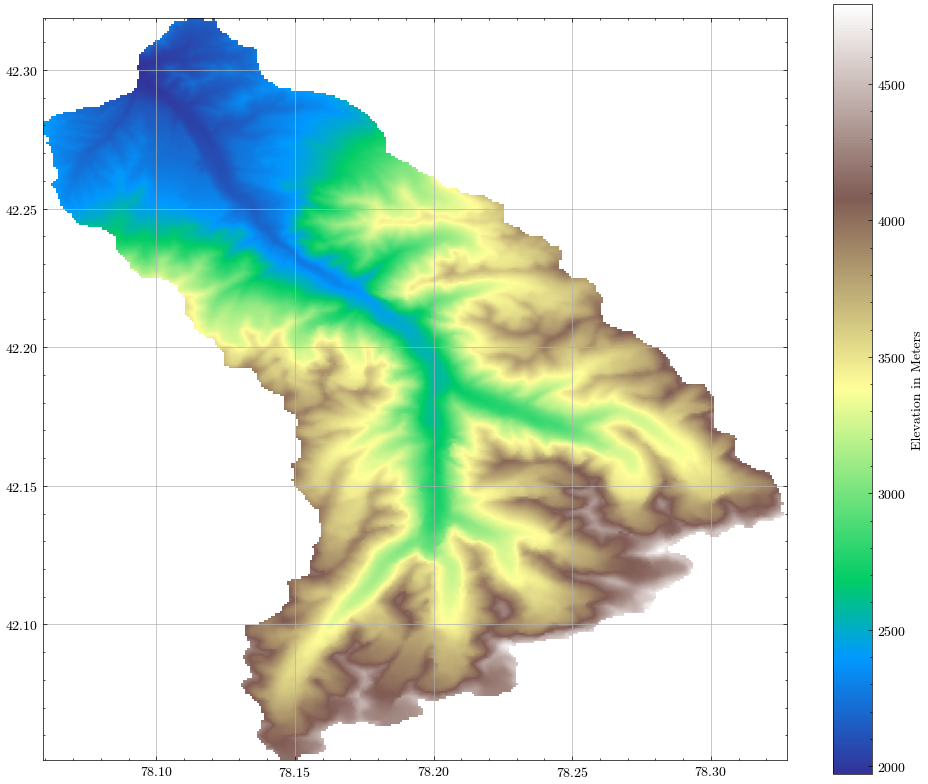
For the following steps, we need the catchment outline in polygon form. Thus, we will convert the raster to a polygon and save both to the output folder in a geopackage. We can calculate the important catchment statistics needed for the glacio-hydrological model in Notebook 4 from these files.
Show code cell source
from shapely.geometry import Polygon, shape
from shapely.ops import transform
import pyproj
# Create shapefile and save it
shapes = grid.polygonize()
schema = {
'geometry': 'Polygon',
'properties': {'LABEL': 'float:16'}
}
catchment_shape = {}
layer_name = 'catchment_orig'
with fiona.open(output_gpkg, 'w',
# driver='ESRI Shapefile',#
driver='GPKG',
layer=layer_name,
crs=grid.crs.srs,
schema=schema) as c:
i = 0
for shape, value in shapes:
catchment_shape = shape
rec = {}
rec['geometry'] = shape
rec['properties'] = {'LABEL': str(value)}
rec['id'] = str(i)
c.write(rec)
i += 1
print(f"Layer '{layer_name}' added to GeoPackage '{output_gpkg}'\n")
catchment_bounds = [int(np.nanmin(demView)), int(np.nanmax(demView))]
ele_cat = float(np.nanmean(demView))
print(f"Catchment elevation ranges from {catchment_bounds[0]} m to {catchment_bounds[1]} m.a.s.l.")
print(f"Mean catchment elevation is {ele_cat:.2f} m.a.s.l.")
Layer 'catchment_orig' added to GeoPackage 'output/catchment_data.gpkg'
Catchment elevation ranges from 1970 m to 4791 m.a.s.l.
Mean catchment elevation is 3274.68 m.a.s.l.
We can also add the catchment polygon to the interactive map. This sends it to GEE and allows us to use a GEE function to calculate its area. Please scroll up to see the results on the map.
Show code cell source
catchment = ee.Geometry.Polygon(catchment_shape['coordinates'])
if show_map:
Map.addLayer(catchment, {}, 'Catchment')
catchment_area = catchment.area().divide(1000 * 1000).getInfo()
print(f"Catchment area is {catchment_area:.2f} km²")
Catchment area is 301.37 km²
Example:
The automatically created box for the pouring point (in gray) is not sufficient to cover the entire catchment area → cropped at the eastern edge.
Manually drawn box (in blue) has been added to ensure that the catchment is not cropped → buffer remains on all edges

Determine glaciers in catchment area#
To acquire outlines of all glaciers in the catchment we will use the Randolph Glacier Inventory version 6 (RGI 6.0). While RGI version 7 has been released, there are no fully compatible ice thickness datasets yet.
The Randolph Glacier Inventory is a global inventory of glacier outlines. It is supplemental to the Global Land Ice Measurements from Space initiative (GLIMS). Production of the RGI was motivated by the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC AR5).
Source: https://www.glims.org/RGI/
The RGI dataset is divided into 19 so called first-order regions.
RGI regions were developed under only three constraints: that they should resemble commonly recognized glacier domains, that together they should contain all of the world’s glaciers, and that their boundaries should be simple and readily recognizable on a map of the world.
Source: Pfeffer et.al. 2014
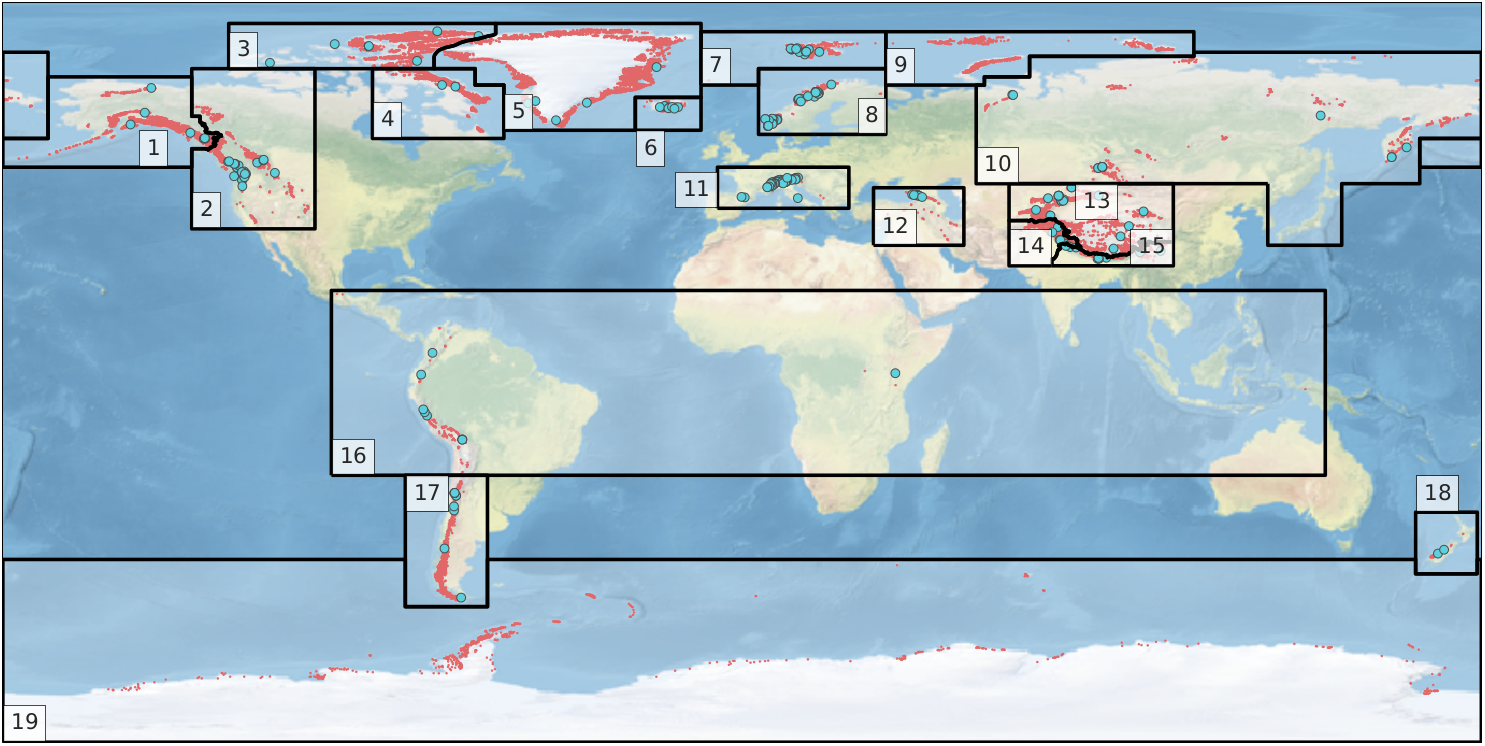
In the first step, the RGI region of the catchment area must be determined to access the correct repository. Therefore, the RGI region outlines will be downloaded and joined with the catchment outline.
Source: RGI Consortium (2017)
Show code cell source
import geopandas as gpd
# load catchment and RGI regions as DF
catchment = gpd.read_file(output_gpkg, layer='catchment_orig')
df_regions = gpd.read_file('https://www.gtn-g.ch/database/GlacReg_2017.zip', layer='GTN-G_glacier_regions_201707')
display(df_regions)
| FULL_NAME | RGI_CODE | WGMS_CODE | geometry | |
|---|---|---|---|---|
| 0 | Alaska | 1 | ALA | POLYGON ((-133 54.5, -134 54.5, -134 54, -134 ... |
| 1 | Alaska | 1 | ALA | POLYGON ((180 50, 179 50, 178 50, 177 50, 176 ... |
| 2 | Western Canada and USA | 2 | WNA | POLYGON ((-133 54.5, -132 54.5, -131 54.5, -13... |
| 3 | Arctic Canada, North | 3 | ACN | POLYGON ((-125 74, -125 75, -125 76, -125 77, ... |
| 4 | Arctic Canada, South | 4 | ACS | POLYGON ((-90 74, -89 74, -88 74, -87 74, -86 ... |
| 5 | Greenland Periphery | 5 | GRL | POLYGON ((-75 77, -74.73 77.51, -74.28 78.06, ... |
| 6 | Iceland | 6 | ISL | POLYGON ((-26 59, -26 60, -26 61, -26 62, -26 ... |
| 7 | Svalbard and Jan Mayen | 7 | SJM | POLYGON ((-10 70, -10 71, -10 72, -10 73, -10 ... |
| 8 | Scandinavia | 8 | SCA | POLYGON ((4 70, 4 71, 4 72, 4 73, 4 74, 5 74, ... |
| 9 | Russian Arctic | 9 | RUA | POLYGON ((35 70, 35 71, 35 72, 35 73, 35 74, 3... |
| 10 | Asia, North | 10 | ASN | POLYGON ((-180 78, -179 78, -178 78, -177 78, ... |
| 11 | Asia, North | 10 | ASN | POLYGON ((128 46, 127 46, 126 46, 125 46, 124 ... |
| 12 | Central Europe | 11 | CEU | POLYGON ((-6 40, -6 41, -6 42, -6 43, -6 44, -... |
| 13 | Caucasus and Middle East | 12 | CAU | POLYGON ((32 31, 32 32, 32 33, 32 34, 32 35, 3... |
| 14 | Asia, Central | 13 | ASC | POLYGON ((80 46, 81 46, 82 46, 83 46, 84 46, 8... |
| 15 | Asia, South West | 14 | ASW | POLYGON ((75.4 26, 75 26, 74 26, 73 26, 72 26,... |
| 16 | Asia, South East | 15 | ASE | POLYGON ((75.4 26, 75.4 27, 75.4 27.62834, 75.... |
| 17 | Low Latitudes | 16 | TRP | POLYGON ((-100 -25, -100 -24, -100 -23, -100 -... |
| 18 | Southern Andes | 17 | SAN | POLYGON ((-62 -45.5, -62 -46, -62 -47, -62 -48... |
| 19 | New Zealand | 18 | NZL | POLYGON ((179 -49, 178 -49, 177 -49, 176 -49, ... |
| 20 | Antarctic and Subantarctic | 19 | ANT | POLYGON ((-180 -45.5, -179 -45.5, -178 -45.5, ... |
For spatial calculations it is crucial to use the correct projection. To avoid inaccuracies due to unit conversions we will project the data to UTM whenever we calculate spatial statistics. The relevant UTM zone and band for the catchment area are determined from the coordinates of the pouring point.
Show code cell source
import utm
from pyproj import CRS
utm_zone = utm.from_latlon(y, x)
print(f"UTM zone '{utm_zone[2]}', band '{utm_zone[3]}'")
# get CRS based on UTM
crs = CRS.from_dict({'proj': 'utm', 'zone': utm_zone[2], 'south': False})
catchment_area = catchment.to_crs(crs).area[0] / 1000 / 1000
print(f"Catchment area (projected) is {catchment_area:.2f} km²")
UTM zone '44', band 'T'
Catchment area (projected) is 302.00 km²
Now we can perform a spatial join between the catchment outline and the RGI regions. If the catchment contains any glaciers, the corresponding RGI region is determined in this step.
Show code cell source
df_regions = df_regions.set_crs('EPSG:4326', allow_override=True)
catchment = catchment.to_crs('EPSG:4326')
df_regions_catchment = gpd.sjoin(df_regions, catchment, how="inner", predicate="intersects")
if len(df_regions_catchment.index) == 0:
print('No area found for catchment')
rgi_region = None
elif len(df_regions_catchment.index) == 1:
rgi_region = df_regions_catchment.iloc[0]['RGI_CODE']
print(f"Catchment belongs to RGI region {rgi_region} ({df_regions_catchment.iloc[0]['FULL_NAME']})")
else:
print("Catchment belongs to more than one region. This use case is not yet supported.")
display(df_regions_catchment)
rgi_region = None
rgi_code = int(df_regions_catchment['RGI_CODE'].iloc[0])
Catchment belongs to RGI region 13 (Asia, Central)
In the next step, the glacier outlines for the determined RGI region will be downloaded. First, we access the repository…
Show code cell source
from resourcespace import ResourceSpace
# use guest credentials to access media server
api_base_url = config['MEDIA_SERVER']['api_base_url']
private_key = config['MEDIA_SERVER']['private_key']
user = config['MEDIA_SERVER']['user']
myrepository = ResourceSpace(api_base_url, user, private_key)
print("Accessed remote repository")
# get resource IDs for each .zip file
rgi_refs = pd.DataFrame(myrepository.get_collection_resources(1168))[
['ref', 'file_size', 'file_extension', 'field8']]
if not rgi_refs.empty:
print("Listing files ...")
display(rgi_refs)
else:
print(f'No files found. Please check remote repository.')
Accessed remote repository
Listing files ...
| ref | file_size | file_extension | field8 | |
|---|---|---|---|---|
| 0 | 27362 | 25220396 | zip | rgi60_17 |
| 1 | 27368 | 15050599 | zip | rgi60_19 |
| 2 | 27358 | 5575712 | zip | rgi60_10 |
| 3 | 27354 | 4525438 | zip | rgi60_08 |
| 4 | 27353 | 20937343 | zip | rgi60_02 |
| 5 | 27352 | 6079435 | zip | rgi60_07 |
| 6 | 27369 | 65663035 | zip | rgi60_13 |
| 7 | 27367 | 82224693 | zip | rgi60_01 |
| 8 | 27366 | 1588336 | zip | rgi60_regions |
| 9 | 27364 | 78735223 | zip | rgi60_05 |
| 10 | 27363 | 46424343 | zip | rgi60_14 |
| 11 | 27361 | 4076485 | zip | rgi60_16 |
| 12 | 27360 | 15918156 | zip | rgi60_15 |
| 13 | 27359 | 5838874 | zip | rgi60_11 |
| 14 | 27357 | 2220673 | zip | rgi60_12 |
| 15 | 27356 | 25777643 | zip | rgi60_04 |
| 16 | 27355 | 4084847 | zip | rgi60_09 |
| 17 | 27350 | 11343491 | zip | rgi60_03 |
| 18 | 27365 | 3050400 | zip | rgi60_18 |
| 19 | 27351 | 2513426 | zip | rgi60_06 |
…and download the .shp files for the target region.
Show code cell source
%%time
import os
import io
from zipfile import ZipFile
output_dir = os.path.join(output_folder, 'RGI')
os.makedirs(output_dir, exist_ok=True)
cnt_rgi = 0
file_names_rgi = []
region_code_str = f'rgi60_{rgi_code:02d}'
#filtering the .zip archives to match our catchment area
filtered_refs = rgi_refs[rgi_refs['field8'] == region_code_str]
if filtered_refs.empty:
print(f'No RGI archive found for region {rgi_code}')
else:
print(f'Found RGI archive(s) for region {rgi_code}:')
display(filtered_refs)
# extracting files from the .zip archives
for _, row in filtered_refs.iterrows():
content = myrepository.get_resource_file(row['ref'], row['file_extension'])
with ZipFile(io.BytesIO(content), 'r') as zipObj:
zipObj.extractall(output_dir)
extracted = zipObj.namelist()
file_names_rgi.extend(extracted)
cnt_rgi += len(extracted)
print(f'{cnt_rgi} files extracted to: {output_dir}')
#reading the shapefile
import glob
region_str = f"{rgi_code}_rgi60_*.shp"
search_pattern = os.path.join(output_folder, 'RGI', region_str)
matching_files = glob.glob(search_pattern)
if matching_files:
rgi_path = matching_files[0]
rgi = gpd.read_file(rgi_path)
print(f"Loaded: {rgi_path}")
else:
print(f"no shapefile for {rgi_code} found")
Found RGI archive(s) for region 13:
| ref | file_size | file_extension | field8 | |
|---|---|---|---|---|
| 6 | 27369 | 65663035 | zip | rgi60_13 |
5 files extracted to: output/RGI
Loaded: output/RGI/13_rgi60_CentralAsia.shp
CPU times: user 1.57 s, sys: 468 ms, total: 2.04 s
Wall time: 27.4 s
Now we can perform a spatial join to determine all glacier outlines that intersect with the catchment area.
Show code cell source
if rgi.crs != catchment.crs:
print("CRS adjusted")
catchment = catchment.to_crs(rgi.crs)
# check whether catchment intersects with glaciers of region
print('Perform spatial join...')
rgi_catchment = gpd.sjoin(rgi, catchment, how='inner', predicate='intersects')
if len(rgi_catchment.index) > 0:
print(f'{len(rgi_catchment.index)} outlines loaded from RGI Region {rgi_code}\n')
Perform spatial join...
51 outlines loaded from RGI Region 13
Some glaciers are not actually in the catchment, but intersect its outline due to spatial inaccuracies. We will first determine their fractional overlap with the target catchment.
Show code cell source
# intersects selects too many. calculate percentage of glacier area that is within catchment
rgi_catchment['rgi_area'] = rgi_catchment.to_crs(crs).area
gdf_joined = gpd.overlay(catchment, rgi_catchment, how='union')
gdf_joined['area_joined'] = gdf_joined.to_crs(crs).area
gdf_joined['share_of_area'] = round((gdf_joined['area_joined'] / gdf_joined['rgi_area'] * 100), 2)
results = (gdf_joined
.groupby(['RGIId', 'LABEL_1'])
.agg({'share_of_area': 'sum'}))
display(results.sort_values(['share_of_area'], ascending=False))
| share_of_area | ||
|---|---|---|
| RGIId | LABEL_1 | |
| RGI60-13.06369 | 1.0 | 100.00 |
| RGI60-13.06368 | 1.0 | 100.00 |
| RGI60-13.07926 | 1.0 | 100.00 |
| RGI60-13.07927 | 1.0 | 100.00 |
| RGI60-13.06381 | 1.0 | 100.00 |
| RGI60-13.06380 | 1.0 | 100.00 |
| RGI60-13.06379 | 1.0 | 100.00 |
| RGI60-13.06376 | 1.0 | 100.00 |
| RGI60-13.06375 | 1.0 | 100.00 |
| RGI60-13.06374 | 1.0 | 100.00 |
| RGI60-13.06373 | 1.0 | 100.00 |
| RGI60-13.06372 | 1.0 | 100.00 |
| RGI60-13.06371 | 1.0 | 100.00 |
| RGI60-13.06370 | 1.0 | 100.00 |
| RGI60-13.07719 | 1.0 | 100.00 |
| RGI60-13.06367 | 1.0 | 100.00 |
| RGI60-13.06361 | 1.0 | 100.00 |
| RGI60-13.06355 | 1.0 | 100.00 |
| RGI60-13.06356 | 1.0 | 100.00 |
| RGI60-13.06357 | 1.0 | 100.00 |
| RGI60-13.06358 | 1.0 | 100.00 |
| RGI60-13.06360 | 1.0 | 100.00 |
| RGI60-13.06362 | 1.0 | 100.00 |
| RGI60-13.06363 | 1.0 | 100.00 |
| RGI60-13.06364 | 1.0 | 100.00 |
| RGI60-13.06365 | 1.0 | 100.00 |
| RGI60-13.06366 | 1.0 | 100.00 |
| RGI60-13.07930 | 1.0 | 99.91 |
| RGI60-13.06377 | 1.0 | 99.80 |
| RGI60-13.07718 | 1.0 | 99.74 |
| RGI60-13.06359 | 1.0 | 99.71 |
| RGI60-13.07931 | 1.0 | 99.44 |
| RGI60-13.06378 | 1.0 | 99.36 |
| RGI60-13.06425 | 1.0 | 98.84 |
| RGI60-13.06382 | 1.0 | 98.11 |
| RGI60-13.07717 | 1.0 | 97.04 |
| RGI60-13.06354 | 1.0 | 90.11 |
| RGI60-13.06353 | 1.0 | 70.49 |
| RGI60-13.06397 | 1.0 | 7.07 |
| RGI60-13.06395 | 1.0 | 5.43 |
| RGI60-13.06451 | 1.0 | 5.01 |
| RGI60-13.07891 | 1.0 | 2.74 |
| RGI60-13.06452 | 1.0 | 2.46 |
| RGI60-13.06421 | 1.0 | 2.44 |
| RGI60-13.06396 | 1.0 | 2.30 |
| RGI60-13.06424 | 1.0 | 1.30 |
| RGI60-13.07925 | 1.0 | 1.01 |
| RGI60-13.06426 | 1.0 | 0.53 |
| RGI60-13.06387 | 1.0 | 0.15 |
| RGI60-13.06386 | 1.0 | 0.10 |
| RGI60-13.06454 | 1.0 | 0.02 |
Now we can filter based on the percentage of shared area. After that the catchment area will be adjusted as follows:
≥50% of the area are in the catchment → include and extend catchment area by full glacier outlines (if needed)
<50% of the area are in the catchment → exclude and reduce catchment area by glacier outlines (if needed)
Show code cell source
rgi_catchment_merge = pd.merge(rgi_catchment, results, on="RGIId")
rgi_in_catchment = rgi_catchment_merge.loc[rgi_catchment_merge['share_of_area'] >= 50]
rgi_out_catchment = rgi_catchment_merge.loc[rgi_catchment_merge['share_of_area'] < 50]
catchment_new = gpd.overlay(catchment, rgi_out_catchment, how='difference')
catchment_new = gpd.overlay(catchment_new, rgi_in_catchment, how='union')
catchment_new = catchment_new.dissolve()[['LABEL_1', 'geometry']]
print(f'Total number of determined glacier outlines: {len(rgi_catchment_merge)}')
print(f'Number of included glacier outlines (overlap >= 50%): {len(rgi_in_catchment)}')
print(f'Number of excluded glacier outlines (overlap < 50%): {len(rgi_out_catchment)}')
Total number of determined glacier outlines: 51
Number of included glacier outlines (overlap >= 50%): 38
Number of excluded glacier outlines (overlap < 50%): 13
The RGI-IDs of the remaining glaciers are stored in Glaciers_in_catchment.csv.
Show code cell source
from pathlib import Path
Path(output_folder + 'RGI').mkdir(parents=True, exist_ok=True)
glacier_ids = pd.DataFrame(rgi_in_catchment)
glacier_ids['RGIId'] = glacier_ids['RGIId'].map(lambda x: str(x).lstrip('RGI60-'))
glacier_ids.to_csv(output_folder + 'RGI/' + 'Glaciers_in_catchment.csv', columns=['RGIId', 'GLIMSId'], index=False)
display(glacier_ids)
| RGIId | GLIMSId | BgnDate | EndDate | CenLon | CenLat | O1Region | O2Region | Area | Zmin | ... | Form | TermType | Surging | Linkages | Name | geometry | index_right | LABEL | rgi_area | share_of_area | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 13.06353 | G078245E42230N | 20020825 | -9999999 | 78.245360 | 42.230246 | 13 | 3 | 0.122 | 3827 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.2473 42.22858, 78.24725 42.22858,... | 0 | 1.0 | 1.224364e+05 | 70.49 |
| 1 | 13.06354 | G078260E42217N | 20020825 | -9999999 | 78.259558 | 42.216524 | 13 | 3 | 0.269 | 3788 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.25962 42.21159, 78.25962 42.21161... | 0 | 1.0 | 2.689387e+05 | 90.11 |
| 2 | 13.06355 | G078253E42212N | 20020825 | -9999999 | 78.252936 | 42.211929 | 13 | 3 | 0.177 | 3640 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.25481 42.2114, 78.25437 42.21124,... | 0 | 1.0 | 1.772685e+05 | 100.00 |
| 3 | 13.06356 | G078242E42208N | 20020825 | -9999999 | 78.241914 | 42.207501 | 13 | 3 | 0.397 | 3665 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.24325 42.20383, 78.24278 42.20365... | 0 | 1.0 | 3.976880e+05 | 100.00 |
| 4 | 13.06357 | G078259E42209N | 20020825 | -9999999 | 78.258704 | 42.209002 | 13 | 3 | 0.053 | 3870 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.2596 42.20812, 78.25958 42.20812,... | 0 | 1.0 | 5.305518e+04 | 100.00 |
| 5 | 13.06358 | G078286E42186N | 20020825 | -9999999 | 78.285595 | 42.185778 | 13 | 3 | 0.082 | 3893 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.28462 42.18393, 78.28461 42.18394... | 0 | 1.0 | 8.178833e+04 | 100.00 |
| 6 | 13.06359 | G078305E42146N | 20020825 | -9999999 | 78.304754 | 42.146142 | 13 | 3 | 4.201 | 3362 | ... | 0 | 0 | 9 | 1 | Karabatkak Glacier | POLYGON ((78.30373 42.14478, 78.30374 42.1449,... | 0 | 1.0 | 4.202605e+06 | 99.71 |
| 7 | 13.06360 | G078285E42147N | 20020825 | -9999999 | 78.284766 | 42.146579 | 13 | 3 | 0.253 | 3689 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.2866 42.14613, 78.2866 42.14613, ... | 0 | 1.0 | 2.530299e+05 | 100.00 |
| 8 | 13.06361 | G078273E42140N | 20020825 | -9999999 | 78.272803 | 42.140310 | 13 | 3 | 2.046 | 3328 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.27549 42.13342, 78.27502 42.13358... | 0 | 1.0 | 2.047230e+06 | 100.00 |
| 9 | 13.06362 | G078260E42141N | 20020825 | -9999999 | 78.260391 | 42.140731 | 13 | 3 | 0.140 | 3912 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.25779 42.1406, 78.25783 42.14063,... | 0 | 1.0 | 1.402120e+05 | 100.00 |
| 10 | 13.06363 | G078257E42156N | 20020825 | -9999999 | 78.257462 | 42.156250 | 13 | 3 | 0.120 | 3586 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.25477 42.15317, 78.25478 42.15319... | 0 | 1.0 | 1.198357e+05 | 100.00 |
| 11 | 13.06364 | G078248E42152N | 20020825 | -9999999 | 78.248267 | 42.151885 | 13 | 3 | 0.488 | 3763 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.24228 42.15276, 78.24179 42.15291... | 0 | 1.0 | 4.878905e+05 | 100.00 |
| 12 | 13.06365 | G078247E42144N | 20020825 | -9999999 | 78.247108 | 42.143715 | 13 | 3 | 0.455 | 3772 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.24121 42.14288, 78.24115 42.14316... | 0 | 1.0 | 4.554036e+05 | 100.00 |
| 13 | 13.06366 | G078256E42146N | 20020825 | -9999999 | 78.256422 | 42.145717 | 13 | 3 | 0.064 | 4091 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.25781 42.14643, 78.25795 42.14627... | 0 | 1.0 | 6.394580e+04 | 100.00 |
| 14 | 13.06367 | G078273E42129N | 20020825 | -9999999 | 78.272638 | 42.128576 | 13 | 3 | 0.838 | 3886 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.27334 42.13093, 78.27335 42.13092... | 0 | 1.0 | 8.380628e+05 | 100.00 |
| 15 | 13.06368 | G078248E42120N | 20020825 | -9999999 | 78.247623 | 42.120056 | 13 | 3 | 0.130 | 3783 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.24502 42.1209, 78.24501 42.12111,... | 0 | 1.0 | 1.303180e+05 | 100.00 |
| 16 | 13.06369 | G078235E42101N | 20020825 | -9999999 | 78.235414 | 42.101281 | 13 | 3 | 1.530 | 3542 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.22665 42.09609, 78.22619 42.09593... | 0 | 1.0 | 1.530797e+06 | 100.00 |
| 17 | 13.06370 | G078176E42094N | 20020825 | -9999999 | 78.175621 | 42.094369 | 13 | 3 | 0.051 | 3715 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.17538 42.09485, 78.17582 42.0949,... | 0 | 1.0 | 5.125522e+04 | 100.00 |
| 18 | 13.06371 | G078177E42081N | 20020825 | -9999999 | 78.177069 | 42.081380 | 13 | 3 | 0.079 | 4111 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.17566 42.08282, 78.17543 42.08271... | 0 | 1.0 | 7.865735e+04 | 100.00 |
| 19 | 13.06372 | G078179E42089N | 20020825 | -9999999 | 78.178560 | 42.088990 | 13 | 3 | 1.188 | 3717 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.18269 42.08684, 78.18287 42.08685... | 0 | 1.0 | 1.189120e+06 | 100.00 |
| 20 | 13.06373 | G078167E42090N | 20020825 | -9999999 | 78.166618 | 42.090111 | 13 | 3 | 0.053 | 3711 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.16486 42.09286, 78.16512 42.09265... | 0 | 1.0 | 5.341084e+04 | 100.00 |
| 21 | 13.06374 | G078164E42077N | 20020825 | -9999999 | 78.163706 | 42.077043 | 13 | 3 | 0.242 | 3821 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.16123 42.07615, 78.16121 42.07647... | 0 | 1.0 | 2.422097e+05 | 100.00 |
| 22 | 13.06375 | G078169E42079N | 20020825 | -9999999 | 78.169405 | 42.078847 | 13 | 3 | 0.210 | 3954 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.16569 42.08133, 78.16569 42.08144... | 0 | 1.0 | 2.103233e+05 | 100.00 |
| 23 | 13.06376 | G078141E42092N | 20020825 | -9999999 | 78.140577 | 42.092421 | 13 | 3 | 0.312 | 3789 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.14342 42.09126, 78.14307 42.0911,... | 0 | 1.0 | 3.125316e+05 | 100.00 |
| 24 | 13.06377 | G078137E42098N | 20020825 | -9999999 | 78.137046 | 42.097894 | 13 | 3 | 0.262 | 3833 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.13993 42.09554, 78.13922 42.09586... | 0 | 1.0 | 2.623176e+05 | 99.80 |
| 25 | 13.06378 | G078142E42102N | 20020825 | -9999999 | 78.142325 | 42.101983 | 13 | 3 | 0.104 | 3917 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.14482 42.10282, 78.14487 42.10251... | 0 | 1.0 | 1.044770e+05 | 99.36 |
| 26 | 13.06379 | G078149E42104N | 20020825 | -9999999 | 78.149436 | 42.104376 | 13 | 3 | 0.202 | 3816 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.14606 42.10367, 78.14607 42.1037,... | 0 | 1.0 | 2.018695e+05 | 100.00 |
| 27 | 13.06380 | G078164E42125N | 20020825 | -9999999 | 78.163621 | 42.125311 | 13 | 3 | 0.557 | 3619 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.16573 42.12234, 78.16524 42.12175... | 0 | 1.0 | 5.575186e+05 | 100.00 |
| 28 | 13.06381 | G078153E42181N | 20020825 | -9999999 | 78.153380 | 42.180904 | 13 | 3 | 0.461 | 3615 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.15813 42.18126, 78.15832 42.18113... | 0 | 1.0 | 4.607761e+05 | 100.00 |
| 29 | 13.06382 | G078144E42185N | 20020825 | -9999999 | 78.144120 | 42.185207 | 13 | 3 | 0.162 | 3664 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.14286 42.18766, 78.14289 42.18785... | 0 | 1.0 | 1.618856e+05 | 98.11 |
| 37 | 13.06425 | G078148E42061N | 20020825 | -9999999 | 78.147941 | 42.060576 | 13 | 3 | 2.110 | 3626 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.13917 42.05947, 78.13898 42.05981... | 0 | 1.0 | 2.111295e+06 | 98.84 |
| 42 | 13.07717 | G078158E42059N | 20020825 | -9999999 | 78.158126 | 42.059389 | 13 | 3 | 0.115 | 4295 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.15995 42.06078, 78.16006 42.06067... | 0 | 1.0 | 1.149899e+05 | 97.04 |
| 43 | 13.07718 | G078157E42064N | 20020825 | -9999999 | 78.156536 | 42.063639 | 13 | 3 | 0.137 | 3996 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.16032 42.06117, 78.16003 42.06088... | 0 | 1.0 | 1.365792e+05 | 99.74 |
| 44 | 13.07719 | G078162E42069N | 20020825 | -9999999 | 78.162144 | 42.069490 | 13 | 3 | 1.541 | 3673 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.17268 42.07346, 78.17306 42.073, ... | 0 | 1.0 | 1.542085e+06 | 100.00 |
| 47 | 13.07926 | G078270E42120N | 20020825 | -9999999 | 78.270069 | 42.119547 | 13 | 3 | 2.825 | 3580 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.28974 42.11692, 78.28933 42.1166,... | 0 | 1.0 | 2.825799e+06 | 100.00 |
| 48 | 13.07927 | G078263E42108N | 20020825 | -9999999 | 78.262621 | 42.108208 | 13 | 3 | 2.422 | 3517 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.28017 42.11207, 78.28018 42.11191... | 0 | 1.0 | 2.423234e+06 | 100.00 |
| 49 | 13.07930 | G078187E42078N | 20020825 | -9999999 | 78.186730 | 42.077626 | 13 | 3 | 3.804 | 3499 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.19163 42.07564, 78.19154 42.07561... | 0 | 1.0 | 3.805955e+06 | 99.91 |
| 50 | 13.07931 | G078212E42082N | 20020825 | -9999999 | 78.212159 | 42.082173 | 13 | 3 | 3.611 | 3397 | ... | 0 | 0 | 9 | 9 | None | POLYGON ((78.20858 42.09121, 78.20871 42.09088... | 0 | 1.0 | 3.612619e+06 | 99.44 |
38 rows × 27 columns
With the updated catchment outline we can now determine the final area of the catchment and the part covered by glaciers.
Show code cell source
catchment_new['area'] = catchment_new.to_crs(crs)['geometry'].area
area_glac = rgi_in_catchment.to_crs(crs)['geometry'].area
area_glac = area_glac.sum() / 1000000
area_cat = catchment_new.iloc[0]['area'] / 1000000
cat_cent = catchment_new.to_crs(crs).centroid
lat = cat_cent.to_crs('EPSG:4326').y[0]
print(f"New catchment area is {area_cat:.2f} km²")
print(f"Glacierized catchment area is {area_glac:.2f} km²")
New catchment area is 300.66 km²
Glacierized catchment area is 31.83 km²
The files just created are added to the existing geopackage…
Show code cell source
rgi_in_catchment.to_file(output_gpkg, layer='rgi_in', driver='GPKG')
print(f"Layer 'rgi_in' added to GeoPackage '{output_gpkg}'")
rgi_out_catchment.to_file(output_gpkg, layer='rgi_out', driver='GPKG')
print(f"Layer 'rgi_out' added to GeoPackage '{output_gpkg}'")
catchment_new.to_file(output_gpkg, layer='catchment_new', driver='GPKG')
print(f"Layer 'catchment_new' added to GeoPackage '{output_gpkg}'")
Layer 'rgi_in' added to GeoPackage 'output/catchment_data.gpkg'
Layer 'rgi_out' added to GeoPackage 'output/catchment_data.gpkg'
Layer 'catchment_new' added to GeoPackage 'output/catchment_data.gpkg'
…and can also be added to the interactive map…
Show code cell source
c_new = geemap.geopandas_to_ee(catchment_new)
rgi = geemap.geopandas_to_ee(rgi_in_catchment)
if show_map:
Map.addLayer(c_new, {'color': 'orange'}, "Catchment New")
Map.addLayer(rgi, {'color': 'white'}, "RGI60")
print('New layers added.')
New layers added.
…or combined in a simple plot.
Show code cell source
fig, ax = plt.subplots()
catchment_new.plot(color='tan', ax=ax)
rgi_in_catchment.plot(color="white", edgecolor="black", ax=ax)
plt.scatter(x, y, facecolor='blue', s=100)
plt.title("Catchment Area with Pouring Point and Glaciers")
plt.savefig(figures_folder + 'NB1_Glaciers_Catchment.png')
plt.show()
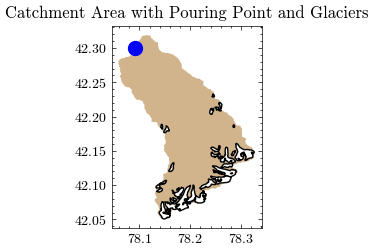
After adding the new catchment area to GEE, we can easily calculate the mean catchment elevation in meters above sea level.
Show code cell source
ele_cat = image.reduceRegion(ee.Reducer.mean(),
geometry=c_new).getInfo()[dem_config[2]]
print(f"Mean catchment elevation (adjusted) is {ele_cat:.2f} m a.s.l.")
Mean catchment elevation (adjusted) is 3271.90 m a.s.l.
Interim Summary:#
So far we have…
…delineated the catchment and determined its area,
…calculated the average elevation of the catchment,
…identified the glaciers in the catchment and calculated their combined area.
In the next step, we will create a glacier profile to determine how the ice is distributed over the elevation range.
Retrieve ice thickness rasters and corresponding DEM files#
Determining ice thickness from remotely sensed data is a challenging task. Fortunately, Farinotti et.al. (2019) calculated an ensemble estimate of different methods for all glaciers in RGI6 and made the data available to the public.
The published repository contains…
(a) the ice thickness distribution of individual glaciers,
(b) global grids at various resolutions with summary-information about glacier number, area, and volume, and
(c) the digital elevation models of the glacier surfaces used to produce the estimates.Nomenclature for glaciers and regions follows the Randolph Glacier Inventory (RGI) version 6.0.
The ice thickness rasters (a) and aligned DEMs (c) are the perfect input data for our glacier profile. The files are selected and downloaded by their RGI IDs and stored in the output folder.
Since the original files hosted by ETH Zurich are stored in large archives, we cut the dataset into smaller slices and reupload them according to the respective license to make them searchable, improve performance, and limit traffic.
First, we identify the relevant archives for our set of glacier IDs.
Show code cell source
def getArchiveNames(row):
region = row['RGIId'].split('.')[0]
id = (int(row['RGIId'].split('.')[1]) - 1) // 1000 + 1
return f'ice_thickness_{region}_{id}', f'dem_surface_DEM_{region}_{id}'
# determine relevant .zip files for derived RGI IDs
df_rgiids = pd.DataFrame(rgi_in_catchment['RGIId'].sort_values())
df_rgiids[['thickness', 'dem']] = df_rgiids.apply(getArchiveNames, axis=1, result_type='expand')
zips_thickness = df_rgiids['thickness'].drop_duplicates()
zips_dem = df_rgiids['dem'].drop_duplicates()
print(f'Thickness archives:\t{zips_thickness.tolist()}')
print(f'DEM archives:\t\t{zips_dem.tolist()}')
Thickness archives: ['ice_thickness_RGI60-13_7', 'ice_thickness_RGI60-13_8']
DEM archives: ['dem_surface_DEM_RGI60-13_7', 'dem_surface_DEM_RGI60-13_8']
The archives are stored on a file server at the Humboldt University of Berlin, which provides limited read access to this notebook. The corresponding credentials and API key are defined in the config.ini file. The next step is to identify the corresponding resource references for the previously identified archives.
Show code cell source
from resourcespace import ResourceSpace
# use guest credentials to access media server
api_base_url = config['MEDIA_SERVER']['api_base_url']
private_key = config['MEDIA_SERVER']['private_key']
user = config['MEDIA_SERVER']['user']
myrepository = ResourceSpace(api_base_url, user, private_key)
# get resource IDs for each .zip file
refs_thickness = pd.DataFrame(myrepository.get_collection_resources(12))[
['ref', 'file_size', 'file_extension', 'field8']]
refs_dem = pd.DataFrame(myrepository.get_collection_resources(21))[['ref', 'file_size', 'file_extension', 'field8']]
# reduce list of resources two required zip files
refs_thickness = pd.merge(zips_thickness, refs_thickness, left_on='thickness', right_on='field8')
refs_dem = pd.merge(zips_dem, refs_dem, left_on='dem', right_on='field8')
print(f'Thickness archive references:\n')
display(refs_thickness)
print(f'DEM archive references:\n')
display(refs_dem)
Thickness archive references:
| thickness | ref | file_size | file_extension | field8 | |
|---|---|---|---|---|---|
| 0 | ice_thickness_RGI60-13_7 | 26948 | 4397086 | zip | ice_thickness_RGI60-13_7 |
| 1 | ice_thickness_RGI60-13_8 | 26986 | 5380873 | zip | ice_thickness_RGI60-13_8 |
DEM archive references:
| dem | ref | file_size | file_extension | field8 | |
|---|---|---|---|---|---|
| 0 | dem_surface_DEM_RGI60-13_7 | 27206 | 8669226 | zip | dem_surface_DEM_RGI60-13_7 |
| 1 | dem_surface_DEM_RGI60-13_8 | 27187 | 11160282 | zip | dem_surface_DEM_RGI60-13_8 |
Again, depending on the number of files and bandwidth, this may take a moment. Let’s start with the ice thickness…
Show code cell source
%%time
import requests
from zipfile import ZipFile
import io
cnt_thickness = 0
file_names_thickness = []
for idx, row in refs_thickness.iterrows():
content = myrepository.get_resource_file(row['ref'], row['file_extension'])
with ZipFile(io.BytesIO(content), 'r') as zipObj:
# Get a list of all archived file names from the zip
listOfFileNames = zipObj.namelist()
for rgiid in df_rgiids.loc[df_rgiids['thickness'] == row['field8']]['RGIId']:
filename = rgiid + '_thickness.tif'
if filename in listOfFileNames:
cnt_thickness += 1
zipObj.extract(filename, output_folder + 'RGI')
file_names_thickness.append(filename)
else:
print(f'File not found: {filename}')
print(f'{cnt_thickness} files have been extracted (ice thickness)')
38 files have been extracted (ice thickness)
CPU times: user 92.5 ms, sys: 54.1 ms, total: 147 ms
Wall time: 3.81 s
…and continue with the matching DEMs.
Show code cell source
%%time
cnt_dem = 0
file_names_dem = []
for idx, row in refs_dem.iterrows():
content = myrepository.get_resource_file(row['ref'])
with ZipFile(io.BytesIO(content), 'r') as zipObj:
# Get a list of all archived file names from the zip
listOfFileNames = zipObj.namelist()
for rgiid in df_rgiids.loc[df_rgiids['dem'] == row['field8']]['RGIId']:
filename = f"surface_DEM_{rgiid}.tif"
if filename in listOfFileNames:
cnt_dem += 1
zipObj.extract(filename, output_folder + 'RGI')
file_names_dem.append(filename)
else:
print(f'File not found: {filename}')
print(f'{cnt_dem} files have been extracted (DEM)')
38 files have been extracted (DEM)
CPU times: user 184 ms, sys: 83.8 ms, total: 268 ms
Wall time: 9.42 s
Show code cell source
if len(rgi_in_catchment) == cnt_thickness == cnt_dem:
print(f"Number of files matches the number of glaciers within catchment: {len(rgi_in_catchment)}")
else:
print("There is a mismatch of extracted files. Please check previous steps for error messages!")
print(f'Number of included glaciers:\t{len(rgi_in_catchment)}')
print(f'Ice thickness files:\t\t{cnt_thickness}')
print(f'DEM files:\t\t\t{cnt_dem}')
Number of files matches the number of glaciers within catchment: 38
Glacier profile creation#
The glacier profile is used to pass the distribution of ice mass in the catchment to the glacio-hydrological model in Notebook 4, following the approach of Seibert et.al.2018. The model then calculates the annual mass balance and redistributes the ice mass accordingly.
To derive the profile from spatially distributed data, we first stack the ice thickness and corresponding DEM rasters for each glacier and create tuples of ice thickness and elevation values.
Show code cell source
from osgeo import gdal
df_all = pd.DataFrame()
if cnt_thickness != cnt_dem:
print('Number of ice thickness raster files does not match number of DEM raster files!')
else:
for idx, rgiid in enumerate(df_rgiids['RGIId']):
if rgiid in file_names_thickness[idx] and rgiid in file_names_dem[idx]:
file_list = [
output_folder + 'RGI/' + file_names_thickness[idx],
output_folder + 'RGI/' + file_names_dem[idx]
]
array_list = []
# Read arrays
for file in file_list:
src = gdal.Open(file)
geotransform = src.GetGeoTransform() # Could be done more elegantly outside the for loop
projection = src.GetProjectionRef()
array_list.append(src.ReadAsArray())
pixelSizeX = geotransform[1]
pixelSizeY = -geotransform[5]
src = None
df = pd.DataFrame()
df['thickness'] = array_list[0].flatten()
df['altitude'] = array_list[1].flatten()
df_all = pd.concat([df_all, df])
else:
print(f'Raster files do not match for {rgiid}')
print("Ice thickness and elevations rasters stacked")
print("Value pairs created")
Ice thickness and elevations rasters stacked
Value pairs created
Now we can remove all data points with zero ice thickness and aggregate all data points into 10m elevation zones. The next step is to calculate the water equivalent (WE) from the average ice thickness in each elevation zone.
The result is exported to the output folder as glacier_profile.csv.
Show code cell source
if len(df_all) > 0:
df_all = df_all.loc[df_all['thickness'] > 0]
df_all.sort_values(by=['altitude'], inplace=True)
# get min/max altitude considering catchment and all glaciers
alt_min = 10 * int(min(catchment_bounds[0], df_all['altitude'].min()) / 10)
alt_max = max(catchment_bounds[1], df_all['altitude'].max()) + 10
# create bins in 10m steps
bins = np.arange(alt_min, df_all['altitude'].max() + 10, 10)
# aggregate per bin and do some math
df_agg = df_all.groupby(pd.cut(df_all['altitude'], bins))['thickness'].agg(count='size', mean='mean').reset_index()
df_agg['Elevation'] = df_agg['altitude'].apply(lambda x: x.left).astype(int)
df_agg['Area'] = df_agg['count'] * pixelSizeX * pixelSizeY / catchment_new.iloc[0]['area']
df_agg['WE'] = df_agg['mean'] * 0.908 * 1000
df_agg['EleZone'] = df_agg['Elevation'].apply(lambda x: 100 * int(x / 100))
# delete empty elevation bands but keep at least one entry per elevation zone
df_agg = pd.concat([df_agg.loc[df_agg['count'] > 0],
df_agg.loc[df_agg['count'] == 0].drop_duplicates(['EleZone'], keep='first')]
).sort_index()
df_agg.drop(['altitude', 'count', 'mean'], axis=1, inplace=True)
df_agg = df_agg.replace(np.nan, 0)
df_agg.to_csv(output_folder + 'glacier_profile.csv', header=True, index=False)
print('Glacier profile for catchment created!\n')
display(df_agg)
Glacier profile for catchment created!
| Elevation | Area | WE | EleZone | |
|---|---|---|---|---|
| 0 | 1970 | 0.000000 | 0.000000 | 1900 |
| 3 | 2000 | 0.000000 | 0.000000 | 2000 |
| 13 | 2100 | 0.000000 | 0.000000 | 2100 |
| 23 | 2200 | 0.000000 | 0.000000 | 2200 |
| 33 | 2300 | 0.000000 | 0.000000 | 2300 |
| ... | ... | ... | ... | ... |
| 276 | 4730 | 0.000023 | 20721.369141 | 4700 |
| 277 | 4740 | 0.000012 | 14450.217773 | 4700 |
| 278 | 4750 | 0.000006 | 10551.472656 | 4700 |
| 279 | 4760 | 0.000000 | 0.000000 | 4700 |
| 281 | 4780 | 0.000002 | 6084.745605 | 4700 |
161 rows × 4 columns
Let’s visualize the glacier profile. First we aggregate the ice mass in larger elevation zones for better visibility. The level of aggregation can be adjusted using the variable steps (default is 20m).
Show code cell source
# aggregation level for plot -> feel free to adjust
steps = 20
# get elevation range where glaciers are present
we_range = df_agg.loc[df_agg['WE'] > 0]['Elevation']
we_range.min() // steps * steps
plt_zones = pd.Series(range(int(we_range.min() // steps * steps),
int(we_range.max() // steps * steps + steps),
steps), name='EleZone').to_frame().set_index('EleZone')
# calculate glacier mass and aggregate glacier profile to defined elevation steps
plt_data = df_agg.copy()
plt_data['EleZone'] = plt_data['Elevation'].apply(lambda x: int(x // steps * steps))
plt_data['Mass'] = plt_data['Area'] * catchment_new.iloc[0]['area'] * plt_data['WE'] * 1e-9 # mass in Mt
plt_data = plt_data.drop(['Area', 'WE'], axis=1).groupby('EleZone').sum().reset_index().set_index('EleZone')
plt_data = plt_zones.join(plt_data)
display(plt_data)
| Elevation | Mass | |
|---|---|---|
| EleZone | ||
| 3320 | 6650 | 0.061096 |
| 3340 | 6690 | 0.485334 |
| 3360 | 6730 | 1.311347 |
| 3380 | 6770 | 2.808075 |
| 3400 | 6810 | 5.413687 |
| ... | ... | ... |
| 4700 | 9410 | 0.410695 |
| 4720 | 9450 | 0.402655 |
| 4740 | 9490 | 0.073972 |
| 4760 | 4760 | 0.000000 |
| 4780 | 4780 | 0.003803 |
74 rows × 2 columns
Now, we can plot the estimated glacier mass (in Mt) for each elevation zone.
Show code cell source
import matplotlib.ticker as ticker
fig, ax = plt.subplots(figsize=(4, 5))
plt_data.plot.barh(y='Mass', ax=ax)
ax.set_xlabel("Glacier mass [Mt]")
ax.set_yticks(ax.get_yticks()[::int(100 / steps)])
ax.set_ylabel("Elevation zone [m a.s.l.]")
ax.get_legend().remove()
plt.title("Initial Ice Distribution")
plt.tight_layout()
plt.savefig(figures_folder + 'NB1_Glacier_Mass_Elevation.png')
plt.show()
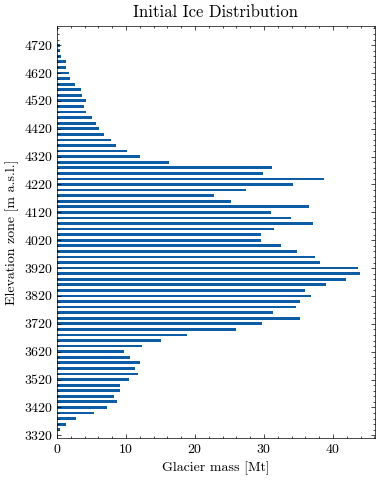
Finally, we calculate the average glacier elevation in meters above sea level.
Show code cell source
ele_glac = round(df_all.altitude.mean(), 2)
print(f'Average glacier elevation in the catchment: {ele_glac:.2f} m a.s.l.')
Average glacier elevation in the catchment: 4001.88 m a.s.l.
Store calculated values for other notebooks#
Create a settings.yml and store the relevant catchment information for the model setup:
area_cat: area of the catchment in km²
ele_cat: average elevation of the catchment in m.a.s.l.
area_glac: glacier covered area as of 2000 in km²
ele_glac: average elevation of glacier covered area in m.a.s.l.
lat: latitude of catchment centroid
Show code cell source
import yaml
settings = {'area_cat': float(area_cat),
'ele_cat': float(ele_cat),
'area_glac': float(area_glac),
'ele_glac': float(ele_glac),
'lat': float(lat)
}
with open(output_folder + 'settings.yml', 'w') as f:
yaml.safe_dump(settings, f)
print('Settings saved to file.')
display(pd.DataFrame(settings.items(), columns=['Parameter', 'Value']).set_index('Parameter'))
Settings saved to file.
| Value | |
|---|---|
| Parameter | |
| area_cat | 300.663718 |
| ele_cat | 3271.895648 |
| area_glac | 31.829413 |
| ele_glac | 4001.879883 |
| lat | 42.185117 |
You can now continue with Notebook 2 or …
Optional: Download Outputs#
output_download.zip). This is especially useful if you want to use the binder environment again, but don't want to start over from Notebook 1.
Show code cell source
import shutil
if zip_output:
shutil.make_archive('output_download', 'zip', 'output')
print('Output folder can be download now (file output_download.zip)')
Output folder can be download now (file output_download.zip)
Show code cell source
%reset -f
